Selection of functional T cell receptor mutants from a yeast surface-display library
- PMID: 10318939
- PMCID: PMC21915
- DOI: 10.1073/pnas.96.10.5651
Selection of functional T cell receptor mutants from a yeast surface-display library
Abstract
The heterodimeric alphabeta T cell receptor (TCR) for antigen is the key determinant of T cell specificity. The structure of the TCR is very similar to that of antibodies, but the engineering of TCRs by directed evolution with combinatorial display libraries has not been accomplished to date. Here, we report that yeast surface display of a TCR was achieved only after the mutation of specific variable region residues. These residues are located in two regions of the TCR, at the interface of the alpha- and beta-chains and in the beta-chain framework region that is thought to be in proximity to the CD3 signal-transduction complex. The mutations are encoded naturally in many antibody variable regions, indicating specific functional differences that have not been appreciated between TCRs and antibodies. The identification of these residues provides an explanation for the inherent difficulties in the display of wild-type TCRs compared with antibodies. Yeast-displayed mutant TCRs bind specifically to the peptide/MHC antigen, enabling engineering of soluble T cell receptors as specific T cell antagonists. This strategy of random mutagenesis followed by selection for surface expression may be of general use in the directed evolution of other eukaryotic proteins that are refractory to display.
Figures
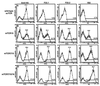

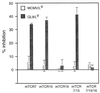
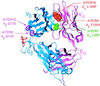
References
-
- Manning T C, Schlueter C J, Brodnicki T C, Parke E A, Speir J A, Garcia K C, Teyton L, Wilson I A, Kranz D M. Immunity. 1998;8:413–425. - PubMed
-
- Alam S M, Travers P J, Wung J L, Nasholds W, Redpath S, Jameson S C, Gascoigne R J. Nature (London) 1996;381:616–620. - PubMed
-
- Garcia K C, Scott C A, Brunmark A, Carbone F R, Peterson P A, Wilson I A, Teyton L. Nature (London) 1996;384:577–581. - PubMed
-
- Bentley G A, Boulot G, Karjalainen K, Mariuzza R A. Science. 1995;267:1984–1987. - PubMed
-
- Fields B A, Ober B, Malchiodi E L, Lebedeva M I, Braden B C, Ysern X, Kim J K, Shao X, Ward E S, Mariuzza R A. Science. 1995;270:1821–1824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

