Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus
- PMID: 10318954
- PMCID: PMC21930
- DOI: 10.1073/pnas.96.10.5740
Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus
Erratum in
- Proc Natl Acad Sci U S A 1999 Aug 31;96(18):10548
Abstract
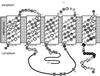
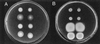
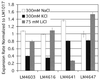
The bacterial flagellum is powered by a rotary motor capable of turning the helical flagellar propeller at very high speeds. Energy to drive rotation is derived from the transmembrane electrochemical potential of specific ions. Ions passing through a channel component are thought to generate the force to power rotation. Two kinds of motors, dependent on different coupling ions, have been described: proton-driven and sodium-driven motors. There are four known genes encoding components of the sodium-powered polar flagellar motor in Vibrio parahaemolyticus. Two, which are characterized here, are homologous to genes encoding constituents of the proton-type motor (motA and motB), and two encode components unique to the sodium-type motor (motX and motY). The sodium-channel-blocking drugs phenamil and amiloride inhibit rotation of the polar flagellum and therefore can be used to probe the architecture of the motor. Mutants were isolated that could swim in the presence of phenamil or amiloride. The majority of the mutations conferring phenamil-resistant motility alter nucleotides in the motA or motB genes. The resultant amino acid changes localize to the cytoplasmic face of the torque generator and permit identification of potential sodium-interaction sites. Mutations that confer motility in the presence of amiloride do not alter any known component of the sodium-type flagellar motor. Thus, evidence supports the existence of more than one class of sodium-interaction site at which inhibitors can interfere with sodium-driven motility.
Figures


Similar articles
-
MotX, the channel component of the sodium-type flagellar motor.J Bacteriol. 1994 Oct;176(19):5988-98. doi: 10.1128/jb.176.19.5988-5998.1994. J Bacteriol. 1994. PMID: 7928960 Free PMC article.
-
Hybrid motor with H(+)- and Na(+)-driven components can rotate Vibrio polar flagella by using sodium ions.J Bacteriol. 1999 Oct;181(20):6332-8. doi: 10.1128/JB.181.20.6332-6338.1999. J Bacteriol. 1999. PMID: 10515922 Free PMC article.
-
Ion-coupling determinants of Na+-driven and H+-driven flagellar motors.J Mol Biol. 2003 Mar 21;327(2):453-63. doi: 10.1016/s0022-2836(03)00096-2. J Mol Biol. 2003. PMID: 12628250
-
Sodium-driven motor of the polar flagellum in marine bacteria Vibrio.Genes Cells. 2011 Oct;16(10):985-99. doi: 10.1111/j.1365-2443.2011.01545.x. Epub 2011 Sep 5. Genes Cells. 2011. PMID: 21895888 Review.
-
Na(+)-driven flagellar motor of Vibrio.Biochim Biophys Acta. 2001 May 1;1505(1):82-93. doi: 10.1016/s0005-2728(00)00279-6. Biochim Biophys Acta. 2001. PMID: 11248191 Review.
Cited by
-
Cysteine-scanning mutagenesis of the periplasmic loop regions of PomA, a putative channel component of the sodium-driven flagellar motor in Vibrio alginolyticus.J Bacteriol. 2000 Feb;182(4):1001-7. doi: 10.1128/JB.182.4.1001-1007.2000. J Bacteriol. 2000. PMID: 10648526 Free PMC article.
-
Novel Amiloride Derivatives That Inhibit Bacterial Motility across Multiple Strains and Stator Types.J Bacteriol. 2021 Oct 25;203(22):e0036721. doi: 10.1128/JB.00367-21. Epub 2021 Sep 13. J Bacteriol. 2021. PMID: 34516280 Free PMC article.
-
Two redundant sodium-driven stator motor proteins are involved in Aeromonas hydrophila polar flagellum rotation.J Bacteriol. 2009 Apr;191(7):2206-17. doi: 10.1128/JB.01526-08. Epub 2009 Jan 30. J Bacteriol. 2009. PMID: 19181813 Free PMC article.
-
Motility of Vibrio spp.: regulation and controlling strategies.Appl Microbiol Biotechnol. 2020 Oct;104(19):8187-8208. doi: 10.1007/s00253-020-10794-7. Epub 2020 Aug 20. Appl Microbiol Biotechnol. 2020. PMID: 32816086 Review.
-
Coupling ion specificity of chimeras between H(+)- and Na(+)-driven motor proteins, MotB and PomB, in Vibrio polar flagella.EMBO J. 2000 Jul 17;19(14):3639-48. doi: 10.1093/emboj/19.14.3639. EMBO J. 2000. PMID: 10899118 Free PMC article.
References
-
- Blair D F. Annu Rev Microbiol. 1995;49:489–522. - PubMed
-
- DeRosier D J. Cell. 1998;93:17–20. - PubMed
-
- Macnab R M. In: Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R, Gross C A, Ingraham J L, Lin E C C, Brooks Low K Jr, Magasanik B, Reznikoff W, Riley M, Schaechter M, et al., editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 123–145.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

