A nod factor binding lectin with apyrase activity from legume roots
- PMID: 10318974
- PMCID: PMC21950
- DOI: 10.1073/pnas.96.10.5856
A nod factor binding lectin with apyrase activity from legume roots
Abstract
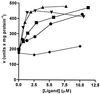
A lectin isolated from the roots of the legume, Dolichos biflorus, binds to Nod factors produced by rhizobial strains that nodulate this plant and has a deduced amino acid sequence with no significant homology to any lectin reported to date. This lectin also is an enzyme that catalyzes the hydrolysis of phosphoanhydride bonds of nucleoside di- and triphosphates; the enzyme activity is increased in the presence of carbohydrate ligands. This lectin-nucleotide phosphohydrolase (LNP) has a substrate specificity characteristic of the apyrase category of phosphohydrolases, and its sequence contains four motifs characteristic of this category of enzymes. LNP is present on the surface of the root hairs, and treatment of roots with antiserum to LNP inhibits their ability to undergo root hair deformation and to form nodules on exposure to rhizobia. These properties suggest that this protein may play a role in the rhizobium-legume symbiosis and/or in a related carbohydrate recognition event endogenous to the plant.
Figures



Similar articles
-
Localization of a Nod factor-binding protein in legume roots and factors influencing its distribution and expression.Plant Physiol. 2000 Nov;124(3):1039-48. doi: 10.1104/pp.124.3.1039. Plant Physiol. 2000. PMID: 11080281 Free PMC article.
-
A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes.Mol Gen Genet. 1999 Sep;262(2):261-7. doi: 10.1007/s004380051082. Mol Gen Genet. 1999. PMID: 10517321
-
Heterologous rhizobial lipochitin oligosaccharides and chitin oligomers induce cortical cell divisions in red clover roots, transformed with the pea lectin gene.Mol Plant Microbe Interact. 2000 Mar;13(3):268-76. doi: 10.1094/MPMI.2000.13.3.268. Mol Plant Microbe Interact. 2000. PMID: 10707352
-
Role of lectins (and rhizobial exopolysaccharides) in legume nodulation.Curr Opin Plant Biol. 1999 Aug;2(4):320-6. doi: 10.1016/S1369-5266(99)80056-9. Curr Opin Plant Biol. 1999. PMID: 10458994 Review.
-
Carbohydrate determinants of Rhizobium-legume symbioses.Carbohydr Res. 1999 Apr 30;317(1-4):1-9. doi: 10.1016/s0008-6215(99)00075-0. Carbohydr Res. 1999. PMID: 10466203 Review.
Cited by
-
Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis.Plant Physiol. 2004 Feb;134(2):595-604. doi: 10.1104/pp.103.031518. Epub 2004 Jan 22. Plant Physiol. 2004. PMID: 14739349 Free PMC article.
-
Expression of the apyrase-like APY1 genes in roots of Medicago truncatula is induced rapidly and transiently by stress and not by Sinorhizobium meliloti or Nod factors.Plant Physiol. 2003 Mar;131(3):1124-36. doi: 10.1104/pp.102.010926. Plant Physiol. 2003. PMID: 12644663 Free PMC article.
-
Chitooligosaccharide sensing and downstream signaling: contrasted outcomes in pathogenic and beneficial plant-microbe interactions.Planta. 2010 Sep;232(4):787-806. doi: 10.1007/s00425-010-1215-9. Epub 2010 Jul 16. Planta. 2010. PMID: 20635098 Review.
-
Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.Protein Sci. 2017 Aug;26(8):1627-1638. doi: 10.1002/pro.3199. Epub 2017 Jun 6. Protein Sci. 2017. PMID: 28543850 Free PMC article.
-
Nitrogen fixation in maize: breeding opportunities.Theor Appl Genet. 2021 May;134(5):1263-1280. doi: 10.1007/s00122-021-03791-5. Epub 2021 Mar 7. Theor Appl Genet. 2021. PMID: 33677701 Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

