Identification and transcriptional control of the genes encoding the Caulobacter crescentus ClpXP protease
- PMID: 10322004
- PMCID: PMC93758
- DOI: 10.1128/JB.181.10.3039-3050.1999
Identification and transcriptional control of the genes encoding the Caulobacter crescentus ClpXP protease
Abstract
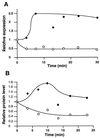
The region of the Caulobacter crescentus chromosome harboring the genes for the ClpXP protease was isolated and characterized. Comparison of the deduced amino acid sequences of the C. crescentus ClpP and ClpX proteins with those of their homologues from several gram-positive and gram-negative bacteria revealed stronger conservation for the ATPase regulatory subunit (ClpX) than for the peptidase subunit (ClpP). The C. crescentus clpX gene was shown by complementation analysis to be functional in Escherichia coli. However, clpX from E. coli was not able to substitute for the essential nature of the clpX gene in C. crescentus. The clpP and clpX genes are separated on the C. crescentus chromosome by an open reading frame pointing in the opposite direction from the clp genes, and transcription of clpP and clpX was found to be uncoupled. clpP is transcribed as a monocistronic unit with a promoter (PP1) located immediately upstream of the 5' end of the gene and a terminator structure following its 3' end. PP1 is under heat shock control and is induced upon entry of the cells into the stationary phase. At least three promoters for clpX (PX1, PX2, and PX3) were mapped in the clpP-clpX intergenic region. In contrast to PP1, the clpX promoters were found to be downregulated after heat shock but were also subject to growth phase control. In addition, the clpP and clpX promoters showed different activity patterns during the cell cycle. Together, these results demonstrate that the genes coding for the peptidase and the regulatory subunits of the ClpXP protease are under independent transcriptional control in C. crescentus. Determination of the numbers of ClpP and ClpX molecules per cell suggested that ClpX is the limiting component compared with ClpP.
Figures




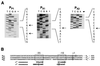


References
-
- Alley M R K, Maddock J R, Shapiro L. Requirement of the carboxyl terminus of the bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1992.
-
- Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
-
- Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

