A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore
- PMID: 10323865
- PMCID: PMC316948
- DOI: 10.1101/gad.13.9.1140
A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore
Abstract
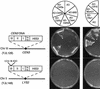
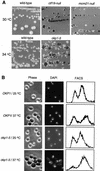
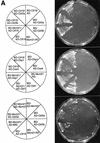
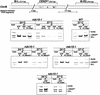
We have established a one-hybrid screen that allows the in vivo localization of proteins at a functional Saccharomyces cerevisiae centromere. Applying this screen we have identified three proteins-Ctf19, Mcm21, and the product of an unspecified open reading frame that we named Okp1-as components of the budding yeast centromere. Ctf19, Mcm21, and Okp1 most likely form a protein complex that links CBF3, a protein complex directly associated with the CDE III element of the centromere DNA, with further components of the budding yeast centromere, Cbf1, Mif2, and Cse4. We demonstrate that the CDE III element is essential and sufficient to localize the established protein network to the centromere and propose that the interaction of the CDE II element with the CDE III localized protein complex facilitates a protein-DNA conformation that evokes the active centromere.
Figures






References
-
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:265–274. - PubMed
-
- Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmids: A uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. - PubMed
-
- Brown MT. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
