Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB)
- PMID: 10325313
- PMCID: PMC84932
- DOI: 10.1128/JCM.37.6.1714-1720.1999
Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB)
Abstract
For the differentiation and identification of mycobacterial species, the rpoB gene, encoding the beta subunit of RNA polymerase, was investigated. rpoB DNAs (342 bp) were amplified from 44 reference strains of mycobacteria and clinical isolates (107 strains) by PCR. The nucleotide sequences were directly determined (306 bp) and aligned by using the multiple alignment algorithm in the MegAlign package (DNASTAR) and the MEGA program. A phylogenetic tree was constructed by the neighbor-joining method. Comparative sequence analysis of rpoB DNAs provided the basis for species differentiation within the genus Mycobacterium. Slowly and rapidly growing groups of mycobacteria were clearly separated, and each mycobacterial species was differentiated as a distinct entity in the phylogenetic tree. Pathogenic Mycobacterium kansasii was easily differentiated from nonpathogenic M. gastri; this differentiation cannot be achieved by using 16S rRNA gene (rDNA) sequences. By being grouped into species-specific clusters with low-level sequence divergence among strains of the same species, all of the clinical isolates could be easily identified. These results suggest that comparative sequence analysis of amplified rpoB DNAs can be used efficiently to identify clinical isolates of mycobacteria in parallel with traditional culture methods and as a supplement to 16S rDNA gene analysis. Furthermore, in the case of M. tuberculosis, rifampin resistance can be simultaneously determined.
Figures


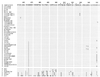
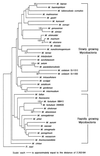
References
-
- Abed Y, Bollet C, de Micco P. Demonstration of Mycobacterium kansasii species heterogeneity by the amplification of the 16S–23S spacer region. J Med Microbiol. 1995;43:156–158. - PubMed
-
- Alekshun M, Kashlev M, Schwartz I. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene. 1997;186:227–235. - PubMed
-
- Bercovier H, Kafri O, Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986;136:1136–1141. - PubMed
-
- Boor K J, Dunkan M L, Price C W. Genetic and transcriptional organization of the region encoding the β subunit of Bacillus subtilis RNA polymerase. J Biol Chem. 1995;270:20329–20336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

