Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization
- PMID: 10325324
- PMCID: PMC84950
- DOI: 10.1128/JCM.37.6.1782-1789.1999
Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization
Abstract
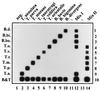
A reverse line blot (RLB) assay was developed for the identification of cattle carrying different species of Theileria and Babesia simultaneously. We included Theileria annulata, T. parva, T. mutans, T. taurotragi, and T. velifera in the assay, as well as parasites belonging to the T. sergenti-T. buffeli-T. orientalis group. The Babesia species included were Babesia bovis, B. bigemina, and B. divergens. The assay employs one set of primers for specific amplification of the rRNA gene V4 hypervariable regions of all Theileria and Babesia species. PCR products obtained from blood samples were hybridized to a membrane onto which nine species-specific oligonucleotides were covalently linked. Cross-reactions were not observed between any of the tested species. No DNA sequences from Bos taurus or other hemoparasites (Trypanosoma species, Cowdria ruminantium, Anaplasma marginale, and Ehrlichia species) were amplified. The sensitivity of the assay was determined at 0.000001% parasitemia, enabling detection of the carrier state of most parasites. Mixed DNAs from five different parasites were correctly identified. Moreover, blood samples from cattle experimentally infected with two different parasites reacted only with the corresponding species-specific oligonucleotides. Finally, RLB was used to screen blood samples collected from carrier cattle in two regions of Spain. T. annulata, T. orientalis, and B. bigemina were identified in these samples. In conclusion, the RLB is a versatile technique for simultaneous detection of all bovine tick-borne protozoan parasites. We recommend its use for integrated epidemiological monitoring of tick-borne disease, since RLB can also be used for screening ticks and can easily be expanded to include additional hemoparasite species.
Figures







References
-
- Allsopp B A, Bayliss H A, Allsopp M T E P, Cavalier-Smith T, Bishop R P, Carrington D M, Sohanpal B, Spooner P. Discrimination between six species of Theileria using oligonucleotide probes which detect small-subunit ribosomal RNA sequences. Parasitology. 1993;107:157–165. - PubMed
-
- Allsopp, P. Unpublished data.
-
- Bishop R, Sohanpal B, Kariuki D P, Young A S, Nene V, Bayliss H, Allsopp B A, Spooner P R, Dolan T T, Morzaria S P. Detection of a carrier state in Theileria parva-infected cattle by the polymerase chain reaction. Parasitology. 1992;104:215–232. - PubMed
-
- Bishop R, Allsopp B, Spooner P, Sohanpal B, Morzaria S, Gobright E. Theileria: improved species discrimination using oligonucleotides derived from large-subunit ribosomal RNA sequences. Exp Parasitol. 1995;80:107–115. - PubMed
-
- Bouattour A, Darghouth M A. First report of Babesia divergens in Tunisia. Vet Parasitol. 1996;63:161–165. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

