Comparative mapping of the region of human chromosome 7 deleted in williams syndrome
- PMID: 10330122
- PMCID: PMC310780
Comparative mapping of the region of human chromosome 7 deleted in williams syndrome
Abstract
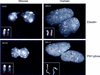
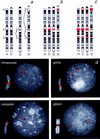
Williams syndrome (WS) is a complex developmental disorder resulting from the deletion of a large (approximately 1.5-2 Mb) segment of human chromosome 7q11.23. Physical mapping studies have revealed that this deleted region, which contains a number of known genes, is flanked by several large, nearly identical blocks of DNA. The presence of such highly related DNA segments in close physical proximity to one another has hampered efforts to elucidate the precise long-range organization of this segment of chromosome 7. To gain insight about the structure and evolutionary origins of this important and complex genomic region, we have constructed a fully contiguous bacterial artificial chromosome (BAC) and P1-derived artificial chromosome (PAC) contig map encompassing the corresponding region on mouse chromosome 5. In contrast to the difficulties encountered in constructing a clone-based physical map of the human WS region, the BAC/PAC-based map of the mouse WS region was straightforward to construct, with no evidence of large duplicated segments, such as those encountered in the human WS region. To confirm this difference, representative human and mouse BACs were used as probes for performing fluorescence in situ hybridization (FISH) to metaphase and interphase chromosomes. Human BACs derived from the nonunique portion of the WS region hybridized to multiple, closely spaced regions on human chromosome 7q11.23. In contrast, corresponding mouse BACs hybridized to a single site on mouse chromosome 5. Furthermore, FISH analysis revealed the presence of duplicated segments within the WS region of various nonhuman primates (chimpanzee, gorilla, orangutan, and gibbon). Hybridization was also noted at the genomic locations corresponding to human chromosome 7p22 and 7q22 in human, chimpanzee, and gorilla, but not in the other animal species examined. Together, these results indicate that the WS region is associated with large, duplicated blocks of DNA on human chromosome 7q11.23 as well as the corresponding genomic regions of other nonhuman primates. However, such duplications are not present in the mouse.
Figures


Similar articles
-
Generation and comparative analysis of approximately 3.3 Mb of mouse genomic sequence orthologous to the region of human chromosome 7q11.23 implicated in Williams syndrome.Genome Res. 2002 Jan;12(1):3-15. doi: 10.1101/gr.214802. Genome Res. 2002. PMID: 11779826 Free PMC article.
-
Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: the low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s).Genomics. 2000 Oct 1;69(1):1-13. doi: 10.1006/geno.2000.6312. Genomics. 2000. PMID: 11013070
-
A complete physical contig and partial transcript map of the Williams syndrome critical region.Genomics. 1999 Jun 1;58(2):138-45. doi: 10.1006/geno.1999.5815. Genomics. 1999. PMID: 10366445
-
Studies on karyotype evolution in higher primates in relation to human chromosome 14 and 9 by comparative mapping of immunoglobulin C epsilon genes with fluorescence in situ hybridization.Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 1999;(117):77-90. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 1999. PMID: 10859938 Review.
-
[Williams syndrome: new insights into genetic etiology, pathogenesis and clinical aspects].Ned Tijdschr Geneeskd. 2001 Mar 3;145(9):396-400. Ned Tijdschr Geneeskd. 2001. PMID: 11253493 Review. Dutch.
Cited by
-
A physical map, including a BAC/PAC clone contig, of the Williams-Beuren syndrome--deletion region at 7q11.23.Am J Hum Genet. 2000 Jan;66(1):47-68. doi: 10.1086/302722. Am J Hum Genet. 2000. PMID: 10631136 Free PMC article.
-
GTF2IRD2 is located in the Williams-Beuren syndrome critical region 7q11.23 and encodes a protein with two TFII-I-like helix-loop-helix repeats.Proc Natl Acad Sci U S A. 2004 Jul 27;101(30):11052-7. doi: 10.1073/pnas.0404150101. Epub 2004 Jul 8. Proc Natl Acad Sci U S A. 2004. PMID: 15243160 Free PMC article.
-
The mosaic structure of human pericentromeric DNA: a strategy for characterizing complex regions of the human genome.Genome Res. 2000 Jun;10(6):839-52. doi: 10.1101/gr.10.6.839. Genome Res. 2000. PMID: 10854415 Free PMC article.
-
Generation and comparative analysis of approximately 3.3 Mb of mouse genomic sequence orthologous to the region of human chromosome 7q11.23 implicated in Williams syndrome.Genome Res. 2002 Jan;12(1):3-15. doi: 10.1101/gr.214802. Genome Res. 2002. PMID: 11779826 Free PMC article.
-
Evolutionary mechanisms shaping the genomic structure of the Williams-Beuren syndrome chromosomal region at human 7q11.23.Genome Res. 2005 Sep;15(9):1179-88. doi: 10.1101/gr.3944605. Genome Res. 2005. PMID: 16140988 Free PMC article.
References
-
- Baumer A, Dutly F, Balmer D, Riegel M, Tukel T, Krajewska-Walasek M, Schinzel AA. High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum Mol Genet. 1998;7:887–894. - PubMed
-
- Bellugi U, Bihrle A, Jernigan T, Trauner D, Doherty S. Neuropsychological, neurological, and neuroanatomical profile of Williams syndrome. Am J Med Genet (Suppl.) 1990;6:115–125. - PubMed
-
- DeBry RW, Seldin MF. Human/mouse homology relationships. Genomics. 1996;33:337–351. - PubMed
-
- Dutly F, Schinzel A. Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams-Beuren syndrome. Hum Mol Genet. 1996;5:1893–1898. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
