Inventory of high-abundance mRNAs in skeletal muscle of normal men
- PMID: 10330131
- PMCID: PMC310786
Inventory of high-abundance mRNAs in skeletal muscle of normal men
Abstract
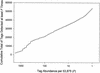
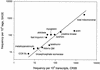
G42875rial analysis of gene expression (SAGE) method was used to generate a catalog of 53,875 short (14 base) expressed sequence tags from polyadenylated RNA obtained from vastus lateralis muscle of healthy young men. Over 12,000 unique tags were detected. The frequency of occurrence of each tag reflects the relative abundance of the corresponding mRNA. The mRNA species that were detected 10 or more times, each comprising >/=0.02% of the mRNA population, accounted for 64% of the mRNA mass but <10% of the total number of mRNA species detected. Almost all of the abundant tags matched mRNA or EST sequences cataloged in GenBank. Mitochondrial transcripts accounted for approximately 20% of the polyadenylated RNA. Transcripts encoding proteins of the myofibrils were the most abundant nuclear-encoded mRNAs. Transcripts encoding ribosomal proteins, and those encoding proteins involved in energy metabolism, also were very abundant. The database can be used as a reference for investigations of alterations in gene expression associated with conditions that influence muscle function, such as muscular dystrophies, aging, and exercise.
Figures



Similar articles
-
High-abundance mRNAs in human muscle: comparison between young and old.J Appl Physiol (1985). 2000 Jul;89(1):297-304. doi: 10.1152/jappl.2000.89.1.297. J Appl Physiol (1985). 2000. PMID: 10904065
-
Transcriptional profile of rat extraocular muscle by serial analysis of gene expression.Invest Ophthalmol Vis Sci. 2002 Apr;43(4):1048-58. Invest Ophthalmol Vis Sci. 2002. PMID: 11923246
-
Pan-genome isolation of low abundance transcripts using SAGE tag.FEBS Lett. 2006 Dec 11;580(28-29):6721-9. doi: 10.1016/j.febslet.2006.11.013. Epub 2006 Nov 14. FEBS Lett. 2006. PMID: 17113583 Free PMC article.
-
Gene expression profile of aging in human muscle.Physiol Genomics. 2003 Jul 7;14(2):149-59. doi: 10.1152/physiolgenomics.00049.2003. Physiol Genomics. 2003. PMID: 12783983
-
[Transcriptomes for serial analysis of gene expression].J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French.
Cited by
-
A comparative analysis of transcript abundance using SAGE and Affymetrix arrays.Funct Integr Genomics. 2005 Jul;5(3):163-74. doi: 10.1007/s10142-005-0135-4. Epub 2005 Feb 16. Funct Integr Genomics. 2005. PMID: 15714318
-
Genetic dissection of platelet function in health and disease using systems biology.Hematol Oncol Clin North Am. 2013 Jun;27(3):443-63. doi: 10.1016/j.hoc.2013.03.002. Epub 2013 Apr 9. Hematol Oncol Clin North Am. 2013. PMID: 23714307 Free PMC article. Review.
-
Proteomic approaches to dissect platelet function: Half the story.Blood. 2006 Dec 15;108(13):3983-91. doi: 10.1182/blood-2006-06-026518. Epub 2006 Aug 22. Blood. 2006. PMID: 16926286 Free PMC article. Review.
-
An anatomy of normal and malignant gene expression.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11287-92. doi: 10.1073/pnas.152324199. Epub 2002 Jul 15. Proc Natl Acad Sci U S A. 2002. PMID: 12119410 Free PMC article.
-
Large functional range of steady-state levels of nuclear and mitochondrial transcripts coding for the subunits of the human mitochondrial OXPHOS system.Genome Res. 2002 Dec;12(12):1901-9. doi: 10.1101/gr.194102. Genome Res. 2002. PMID: 12466294 Free PMC article.
References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. - PubMed
-
- Clayton DA. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. - PubMed
-
- Forsberg AM, Nillson E, Wernerman J, Bergstrom J, Hultman E. Muscle composition in relation to age and sex. Clin Sci. 1991;81:249–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
