The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain
- PMID: 10330133
- PMCID: PMC104352
- DOI: 10.1128/MCB.19.6.3931
The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain
Abstract
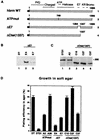
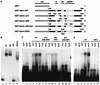
The mammalian SWI-SNF complex is a chromatin-remodelling machinery involved in the modulation of gene expression. Its activity relies on two closely related ATPases known as brm/SNF2alpha and BRG-1/SNF2beta. These two proteins can cooperate with nuclear receptors for transcriptional activation. In addition, they are involved in the control of cell proliferation, most probably by facilitating p105(Rb) repression of E2F transcriptional activity. In the present study, we have examined the ability of various brm/SNF2alpha deletion mutants to reverse the transformed phenotype of ras-transformed fibroblasts. Deletions within the p105(Rb) LXCXE binding motif or the conserved bromodomain had only a moderate effect. On the other hand, a 49-amino-acid segment, rich in lysines and arginines and located immediately downstream of the p105(Rb) interaction domain, appeared to be essential in this assay. This region was also required for cooperation of brm/SNF2alpha with the glucocorticoid receptor in transfection experiments, but only in the context of a reporter construct integrated in the cellular genome. The region has homology to the AT hooks present in high-mobility-group protein I/Y DNA binding domains and is required for the tethering of brm/SNF2alpha to chromatin.
Figures




References
-
- Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. - PubMed
-
- Armstrong J A, Bieker J J, Emerson B M. A Swi/Snf-related chromatin remodeling complex, E-Rc1, is required for tissue-specific transcriptional regulation by Eklf in vitro. Cell. 1998;95:93–104. - PubMed
-
- Ashley C T, Pendleton C G, Jennings W W, Saxena A, Glover C V. Isolation and sequencing of cDNA clones encoding Drosophila chromosomal protein D1. A repeating motif in proteins which recognize at DNA. J Biol Chem. 1989;264:8394–8401. - PubMed
-
- Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. - PubMed
-
- Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
