Dimeric RFX proteins contribute to the activity and lineage specificity of the interleukin-5 receptor alpha promoter through activation and repression domains
- PMID: 10330134
- PMCID: PMC104353
- DOI: 10.1128/MCB.19.6.3940
Dimeric RFX proteins contribute to the activity and lineage specificity of the interleukin-5 receptor alpha promoter through activation and repression domains
Abstract
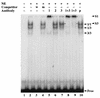
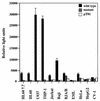
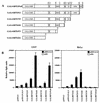
Interleukin-5 (IL-5) plays a central role in the differentiation, proliferation, and functional activation of eosinophils. The specific action of IL-5 on eosinophils and hematopoietically related basophils is regulated by the restricted expression of IL-5 receptor alpha (IL-5Ralpha), a subunit of high-affinity IL-5R, on these cells. We have previously identified an enhancer-like cis element in the IL-5Ralpha promoter that is important for both full promoter function and lineage-specific activity. Here, we demonstrate by yeast one-hybrid screening that RFX2 protein specifically binds to this cis element. RFX2 belongs to the RFX DNA-binding protein family, the biological role of which remains obscure. Using an electrophoretic mobility shift assay, we further show that RFX1, RFX2, and RFX3 homodimers and heterodimers specifically bind to the cis element of the IL-5Ralpha promoter. The mRNA expression of RFX1, RFX2, and RFX3 was detected ubiquitously, but in transient-transfection assays, multimerized RFX binding sites in front of a basal promoter efficiently functioned in a tissue- and lineage-specific manner. To further investigate RFX functions on transcription, full-length and deletion mutants of RFX1 were targeted to DNA through fusion to the GAL4 DNA binding domain. Tissue- and lineage-specific transcriptional activation with the full-length RFX1 fusion plasmid on a reporter controlled by GAL4 binding sites was observed. Distinct activation and repression domains within the RFX1 protein were further mapped. Our findings suggest that RFX proteins are transcription factors that contribute to the activity and lineage specificity of the IL-5Ralpha promoter by directly binding to a target cis element and cooperating with other tissue- and lineage-specific cofactors.
Figures






Similar articles
-
Human regulatory factor X 4 (RFX4) is a testis-specific dimeric DNA-binding protein that cooperates with other human RFX members.J Biol Chem. 2002 Jan 4;277(1):836-42. doi: 10.1074/jbc.M108638200. Epub 2001 Oct 26. J Biol Chem. 2002. PMID: 11682486
-
The dimerization/repression domain of RFX1 is related to a conserved region of its yeast homologues Crt1 and Sak1: a new function for an ancient motif.J Mol Biol. 1999 Nov 19;294(1):121-37. doi: 10.1006/jmbi.1999.3245. J Mol Biol. 1999. PMID: 10556033
-
RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins.Mol Cell Biol. 1994 Feb;14(2):1230-44. doi: 10.1128/mcb.14.2.1230-1244.1994. Mol Cell Biol. 1994. PMID: 8289803 Free PMC article.
-
Involvement of RFX proteins in transcriptional activation from a Ras-responsive enhancer element.Arch Dermatol Res. 2004 Apr;295(11):482-9. doi: 10.1007/s00403-004-0456-5. Epub 2004 Mar 13. Arch Dermatol Res. 2004. PMID: 15024578
-
Interleukin-5 receptor alpha subunit gene regulation in human eosinophil development: identification of a unique cis-element that acts lie an enhancer in regulating activity of the IL-5R alpha promoter.Curr Top Microbiol Immunol. 1996;211:173-87. doi: 10.1007/978-3-642-85232-9_18. Curr Top Microbiol Immunol. 1996. PMID: 8585949 Review.
Cited by
-
Forkhead transcription factor Fd3F cooperates with Rfx to regulate a gene expression program for mechanosensory cilia specialization.Dev Cell. 2012 Jun 12;22(6):1221-33. doi: 10.1016/j.devcel.2012.05.010. Dev Cell. 2012. PMID: 22698283 Free PMC article.
-
Knockout of the regulatory factor X1 gene leads to early embryonic lethality.Biochem Biophys Res Commun. 2009 Sep 4;386(4):715-7. doi: 10.1016/j.bbrc.2009.06.111. Epub 2009 Jun 25. Biochem Biophys Res Commun. 2009. PMID: 19559676 Free PMC article.
-
T cell reactivity to regulatory factor X4 in type 1 narcolepsy.Sci Rep. 2021 Apr 9;11(1):7841. doi: 10.1038/s41598-021-87481-8. Sci Rep. 2021. PMID: 33837283 Free PMC article.
-
The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5.J Virol. 2000 Nov;74(22):10458-67. doi: 10.1128/jvi.74.22.10458-10467.2000. J Virol. 2000. PMID: 11044090 Free PMC article.
-
ZNF143 protein is an important regulator of the myeloid transcription factor C/EBPα.J Biol Chem. 2017 Nov 17;292(46):18924-18936. doi: 10.1074/jbc.M117.811109. Epub 2017 Sep 12. J Biol Chem. 2017. PMID: 28900037 Free PMC article.
References
-
- Baltus B, van Dijk T B, Caldenhoven E, Zanders E, Raaijmakers J A M, Lammers J-W J, Koenderman L, de Groot R P. An AP-1 site in the promoter of the human IL-5Rα gene is necessary for promoter activity in eosinophilic HL60 cells. FEBS Lett. 1998;434:251–254. - PubMed
-
- Chen H M, Zhang P, Voso M T, Hohaus S, Gonzalez D A, Christopher K G, Zhang D-E, Tenen D G. Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood. 1995;85:2918–2928. - PubMed
-
- Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources rich in ribonuclease. Biochemistry. 1979;18:5294–5299. - PubMed
-
- Clausen B E, Waldburger J-M, Schwenk F, Barras E, Mach B, Rajewsky K, Förster I, Reith W. Residual MHC class II expression on mature dendritic cells and activated B cells in RFX5-deficient mice. Immunity. 1998;8:143–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
