A TATA-binding protein mutant defective for TFIID complex formation in vivo
- PMID: 10330135
- PMCID: PMC104354
- DOI: 10.1128/MCB.19.6.3951
A TATA-binding protein mutant defective for TFIID complex formation in vivo
Abstract
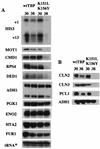
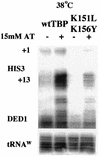
Using an intragenic complementation screen, we have identified a temperature-sensitive TATA-binding protein (TBP) mutant (K151L, K156Y) that is defective for interaction with certain yeast TBP-associated factors (TAFs) at the restrictive temperature. The K151L,K156Y mutant appears to be functional for RNA polymerase I (Pol I) and Pol III transcription, and it is capable of supporting Gal4-activated and Gcn4-activated transcription by Pol II. However, transcription from certain TATA-containing and TATA-less Pol II promoters is reduced at the restrictive temperature. Immunoprecipitation analysis of extracts prepared after culturing cells at the restrictive temperature for 1 h indicates that the K151L,K156Y derivative is severely compromised in its ability to interact with TAF130, TAF90, TAF68/61, and TAF25 while remaining functional for interaction with TAF60 and TAF30. Thus, a TBP mutant that is compromised in its ability to form TFIID can support the response to Gcn4 but is defective for transcription from specific promoters in vivo.
Figures




Similar articles
-
Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAF(II)130.Mol Cell Biol. 1997 Jun;17(6):3081-93. doi: 10.1128/MCB.17.6.3081. Mol Cell Biol. 1997. PMID: 9154807 Free PMC article.
-
Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding.Proc Natl Acad Sci U S A. 1995 Dec 5;92(25):11864-8. doi: 10.1073/pnas.92.25.11864. Proc Natl Acad Sci U S A. 1995. PMID: 8524864 Free PMC article.
-
TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo.Science. 2000 May 19;288(5469):1244-8. doi: 10.1126/science.288.5469.1244. Science. 2000. PMID: 10818000
-
Mechanisms of transcriptional activation and repression can both involve TFIID.Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):517-26. doi: 10.1098/rstb.1996.0050. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735274 Review.
-
Selective roles for TATA-binding-protein-associated factors in vivo.Genes Funct. 1997 Feb;1(1):5-9. doi: 10.1046/j.1365-4624.1997.00004.x. Genes Funct. 1997. PMID: 9680324 Review.
Cited by
-
Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast.Genetics. 2000 Jul;155(3):1045-54. doi: 10.1093/genetics/155.3.1045. Genetics. 2000. PMID: 10880468 Free PMC article.
-
Autonomous function of the amino-terminal inhibitory domain of TAF1 in transcriptional regulation.Mol Cell Biol. 2004 Apr;24(8):3089-99. doi: 10.1128/MCB.24.8.3089-3099.2004. Mol Cell Biol. 2004. PMID: 15060133 Free PMC article.
-
Evidence that TAF-TATA box-binding protein interactions are required for activated transcription in mammalian cells.Mol Cell Biol. 2002 Apr;22(8):2788-98. doi: 10.1128/MCB.22.8.2788-2798.2002. Mol Cell Biol. 2002. PMID: 11909971 Free PMC article.
-
Region of yeast TAF 130 required for TFIID to associate with promoters.Mol Cell Biol. 2001 Feb;21(4):1145-54. doi: 10.1128/MCB.21.4.1145-1154.2001. Mol Cell Biol. 2001. PMID: 11158301 Free PMC article.
-
Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene.Mol Cell Biol. 2000 Apr;20(7):2385-99. doi: 10.1128/MCB.20.7.2385-2399.2000. Mol Cell Biol. 2000. PMID: 10713163 Free PMC article.
References
-
- Apone L M, Virbasius C-A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. - PubMed
-
- Apone L M, Virbasius C A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. - PubMed
-
- Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
