Mitochondrial group II introns, cytochrome c oxidase, and senescence in Podospora anserina
- PMID: 10330149
- PMCID: PMC104368
- DOI: 10.1128/MCB.19.6.4093
Mitochondrial group II introns, cytochrome c oxidase, and senescence in Podospora anserina
Abstract
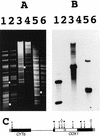
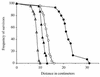
Podospora anserina is a filamentous fungus with a limited life span. It expresses a degenerative syndrome called senescence, which is always associated with the accumulation of circular molecules (senDNAs) containing specific regions of the mitochondrial chromosome. A mobile group II intron (alpha) has been thought to play a prominent role in this syndrome. Intron alpha is the first intron of the cytochrome c oxidase subunit I gene (COX1). Mitochondrial mutants that escape the senescence process are missing this intron, as well as the first exon of the COX1 gene. We describe here the first mutant of P. anserina that has the alpha sequence precisely deleted and whose cytochrome c oxidase activity is identical to that of wild-type cells. The integration site of the intron is slightly modified, and this change prevents efficient homing of intron alpha. We show here that this mutant displays a senescence syndrome similar to that of the wild type and that its life span is increased about twofold. The introduction of a related group II intron into the mitochondrial genome of the mutant does not restore the wild-type life span. These data clearly demonstrate that intron alpha is not the specific senescence factor but rather an accelerator or amplifier of the senescence process. They emphasize the role that intron alpha plays in the instability of the mitochondrial chromosome and the link between this instability and longevity. Our results strongly support the idea that in Podospora, "immortality" can be acquired not by the absence of intron alpha but rather by the lack of active cytochrome c oxidase.
Figures




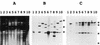
Similar articles
-
DNA deletion of mitochondrial introns is correlated with the process of senescence in Podospora anserina.J Mol Biol. 1993 Nov 5;234(1):1-7. doi: 10.1006/jmbi.1993.1558. J Mol Biol. 1993. PMID: 8230190
-
DNA double-strand break in vivo at the 3' extremity of exons located upstream of group II introns. Senescence and circular DNA introns in Podospora mitochondria.J Mol Biol. 1994 Oct 7;242(5):630-43. doi: 10.1006/jmbi.1994.1613. J Mol Biol. 1994. PMID: 7932720
-
Self-splicing of the mobile group II intron of the filamentous fungus Podospora anserina (COI I1) in vitro.EMBO J. 1990 Jul;9(7):2289-98. doi: 10.1002/j.1460-2075.1990.tb07400.x. EMBO J. 1990. PMID: 2162769 Free PMC article.
-
Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina.Biochim Biophys Acta. 2006 May-Jun;1757(5-6):604-10. doi: 10.1016/j.bbabio.2006.03.005. Epub 2006 Mar 30. Biochim Biophys Acta. 2006. PMID: 16624249 Review.
-
Mitochondrial oxidative stress and aging in the filamentous fungus Podospora anserina.Ann N Y Acad Sci. 2000 Jun;908:31-9. doi: 10.1111/j.1749-6632.2000.tb06633.x. Ann N Y Acad Sci. 2000. PMID: 10911945 Review.
Cited by
-
The Ll.LtrB intron from Lactococcus lactis excises as circles in vivo: insights into the group II intron circularization pathway.RNA. 2015 Jul;21(7):1286-93. doi: 10.1261/rna.046367.114. Epub 2015 May 8. RNA. 2015. PMID: 25956521 Free PMC article.
-
Genetic, parental and lifestyle factors influence telomere length.Commun Biol. 2022 Jun 9;5(1):565. doi: 10.1038/s42003-022-03521-7. Commun Biol. 2022. PMID: 35681050 Free PMC article.
-
A causal link between respiration and senescence in Podospora anserina.Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):4138-43. doi: 10.1073/pnas.070501997. Proc Natl Acad Sci U S A. 2000. PMID: 10759557 Free PMC article.
-
The absence of a mitochondrial genome in rho0 yeast cells extends lifespan independently of retrograde regulation.Exp Gerontol. 2009 Jun-Jul;44(6-7):390-7. doi: 10.1016/j.exger.2009.03.001. Epub 2009 Mar 12. Exp Gerontol. 2009. PMID: 19285548 Free PMC article.
-
The conserved mitochondrial gene distribution in relatives of Turritopsis nutricula, an immortal jellyfish.Bioinformation. 2014 Sep 30;10(9):586-91. doi: 10.6026/97320630010586. eCollection 2014. Bioinformation. 2014. PMID: 25352727 Free PMC article.
References
-
- Belcour L, Begel O. Life-span and senescence in Podospora anserina: effect of mitochondrial genes and functions. J Gen Microbiol. 1980;119:505–515.
-
- Belcour L, Rossignol M, Koll F, Sellem C H, Oldani C. Plasticity of the mitochondrial genome in Podospora. Polymorphism for 15 optional sequences: group-I, group-II introns, intronic ORFs and an intergenic region. Curr Genet. 1997;31:308–317. - PubMed
-
- Belcour L, Sainsard-Chanet A, Sellem C H. Mobile group II introns, DNA circles, reverse transcriptase and senescence. Genetica. 1994;93:225–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
