GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae
- PMID: 10330157
- PMCID: PMC104376
- DOI: 10.1128/MCB.19.6.4167
GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae
Abstract
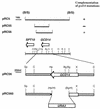
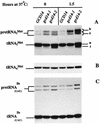
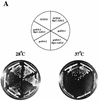
Gcd10p and Gcd14p were first identified genetically as repressors of GCN4 mRNA translation in Saccharomyces cerevisiae. Recent findings indicate that Gcd10p and Gcd14p reside in a nuclear complex required for the presence of 1-methyladenosine in tRNAs. Here we show that Gcd14p is an essential protein with predicted binding motifs for S-adenosylmethionine, consistent with a direct function in tRNA methylation. Two different gcd14 mutants exhibit defects in cell growth and accumulate high levels of initiator methionyl-tRNA (tRNAiMet) precursors containing 5' and 3' extensions, suggesting a defect in processing of the primary transcript. Dosage suppressors of gcd10 mutations, encoding tRNAiMet (hcIMT1 to hcIMT4; hc indicates that the gene is carried on a high-copy-number plasmid) or a homologue of human La protein implicated in tRNA 3'-end formation (hcLHP1), also suppressed gcd14 mutations. In fact, the lethality of a GCD14 deletion was suppressed by hcIMT4, indicating that the essential function of Gcd14p is required for biogenesis of tRNAiMet. A mutation in GCD10 or deletion of LHP1 exacerbated the defects in cell growth and expression of mature tRNAiMet in gcd14 mutants, consistent with functional interactions between Gcd14p, Gcd10p, and Lhp1p in vivo. Surprisingly, the amounts of NME1 and RPR1, the RNA components of RNases P and MRP, were substantially lower in gcd14 lhp1::LEU2 double mutants than in the corresponding single mutants, whereas 5S rRNA was present at wild-type levels. Our findings suggest that Gcd14p and Lhp1p cooperate in the maturation of a subset of RNA polymerase III transcripts.
Figures








Similar articles
-
The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA.Genes Dev. 1998 Dec 1;12(23):3650-62. doi: 10.1101/gad.12.23.3650. Genes Dev. 1998. PMID: 9851972 Free PMC article.
-
The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2000 May 9;97(10):5173-8. doi: 10.1073/pnas.090102597. Proc Natl Acad Sci U S A. 2000. PMID: 10779558 Free PMC article.
-
Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae.Genetics. 1998 Mar;148(3):1007-20. doi: 10.1093/genetics/148.3.1007. Genetics. 1998. PMID: 9539420 Free PMC article.
-
Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2.Mol Cell Biol. 2000 Apr;20(7):2505-16. doi: 10.1128/MCB.20.7.2505-2516.2000. Mol Cell Biol. 2000. PMID: 10713174 Free PMC article.
-
GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3.Genes Dev. 1995 Jul 15;9(14):1781-96. doi: 10.1101/gad.9.14.1781. Genes Dev. 1995. PMID: 7542616
Cited by
-
La protein and the trafficking of nascent RNA polymerase iii transcripts.J Cell Biol. 2001 May 14;153(4):F13-8. doi: 10.1083/jcb.153.4.f13. J Cell Biol. 2001. PMID: 11352926 Free PMC article. Review. No abstract available.
-
Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae.Genes Dev. 2004 Jun 1;18(11):1227-40. doi: 10.1101/gad.1183804. Epub 2004 May 14. Genes Dev. 2004. PMID: 15145828 Free PMC article.
-
Modulation of yeast genome expression in response to defective RNA polymerase III-dependent transcription.Mol Cell Biol. 2005 Oct;25(19):8631-42. doi: 10.1128/MCB.25.19.8631-8642.2005. Mol Cell Biol. 2005. PMID: 16166643 Free PMC article.
-
Precursor-product discrimination by La protein during tRNA metabolism.Nat Struct Mol Biol. 2009 Apr;16(4):430-7. doi: 10.1038/nsmb.1573. Epub 2009 Mar 15. Nat Struct Mol Biol. 2009. PMID: 19287396 Free PMC article.
-
Integrity of SRP RNA is ensured by La and the nuclear RNA quality control machinery.Nucleic Acids Res. 2014;42(16):10698-710. doi: 10.1093/nar/gku761. Epub 2014 Aug 26. Nucleic Acids Res. 2014. PMID: 25159613 Free PMC article.
References
-
- Altschul S F, Boguski M S, Gish W, Wootton J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. - PubMed
-
- Anderson, J., and A. G. Hinnebusch. Unpublished observations.
-
- Asano K, Phan L, Anderson J, Hinnebusch A G. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
