B-Cell coactivator OBF-1 exhibits unusual transcriptional properties and functions in a DNA-bound Oct-1-dependent fashion
- PMID: 10330165
- PMCID: PMC104384
- DOI: 10.1128/MCB.19.6.4247
B-Cell coactivator OBF-1 exhibits unusual transcriptional properties and functions in a DNA-bound Oct-1-dependent fashion
Abstract
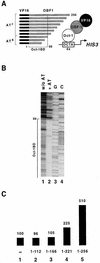
Eukaryotic transcriptional activators generally comprise both a DNA-binding domain that recognizes specific cis-regulatory elements in the target genes and an activation domain which is essential for transcriptional stimulation. Activation domains typically behave as structurally and functionally autonomous modules that retain their intrinsic activities when directed to a promoter by a variety of heterologous DNA-binding domains. Here we report that OBF-1, a B-cell-specific coactivator for transcription factor Oct-1, challenges this traditional view in that it contains an atypical activation domain that exhibits two unexpected functional properties when tested in the yeast Saccharomyces cerevisiae. First, OBF-1 by itself has essentially no intrinsic activation potential, yet it strongly synergizes with other activation domains such as VP16 and Gal4. Second, OBF-1 exerts its effect in association with DNA-bound Oct-1 but is inactive when attached to a heterologous DNA-binding domain. These findings suggest that activation by OBF-1 is not obtained by simple recruitment of the coactivator to the promoter but requires interaction with DNA-bound Oct-1 to stimulate a step distinct from those regulated by classical activation domains.
Figures



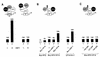
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
