Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA
- PMID: 10330172
- PMCID: PMC104391
- DOI: 10.1128/MCB.19.6.4311
Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA
Abstract
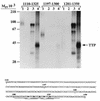
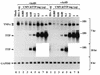
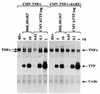
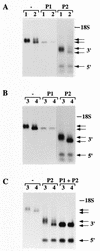
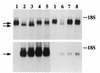
Mice deficient in tristetraprolin (TTP), the prototype of a family of CCCH zinc finger proteins, develop an inflammatory syndrome mediated by excess tumor necrosis factor alpha (TNF-alpha). Macrophages derived from these mice oversecrete TNF-alpha, by a mechanism that involves stabilization of TNF-alpha mRNA, and TTP can bind directly to the AU-rich element (ARE) in TNF-alpha mRNA (E. Carballo, W. S. Lai, and P. J. Blackshear, Science 281:1001-1005, 1998). We show here that TTP binding to the TNF-alpha ARE is dependent upon the integrity of both zinc fingers, since mutation of a single cysteine residue in either zinc finger to arginine severely attenuated the binding of TTP to the TNF-alpha ARE. In intact cells, TTP at low expression levels promoted a decrease in size of the TNF-alpha mRNA as well as a decrease in its amount; at higher expression levels, the shift to a smaller TNF-alpha mRNA size persisted, while the accumulation of this smaller species increased. RNase H experiments indicated that the shift to a smaller size was due to TTP-promoted deadenylation of TNF-alpha mRNA. This CCCH protein is likely to be important in the deadenylation and degradation of TNF-alpha mRNA and perhaps other ARE-containing mRNAs, both in normal physiology and in certain pathological conditions.
Figures

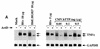
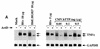


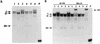
References
-
- Akashi M, Shaw G, Gross M, Saito M, Koeffler H P. Role of AUUUA sequences in stabilization of granulocyte-macrophage colony-stimulating factor RNA in stimulated cells. Blood. 1991;78:2005–2012. - PubMed
-
- Blackshear P J. Systems for polyacrylamide gel electrophoresis. Methods Enzymol. 1984;104:237–255. - PubMed
-
- Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992;267:6302–6309. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
