Collagenase 3 is a target of Cbfa1, a transcription factor of the runt gene family involved in bone formation
- PMID: 10330183
- PMCID: PMC104402
- DOI: 10.1128/MCB.19.6.4431
Collagenase 3 is a target of Cbfa1, a transcription factor of the runt gene family involved in bone formation
Abstract
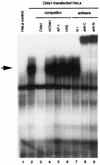
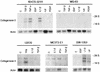
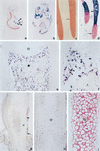
Collagenase 3 (MMP-13) is a recently identified member of the matrix metalloproteinase (MMP) gene family that is expressed at high levels in diverse human carcinomas and in articular cartilage from arthritic patients. In addition to its expression in pathological conditions, collagenase 3 has been detected in osteoblasts and hypertrophic chondrocytes during fetal ossification. In this work, we have evaluated the possibility that Cbfa1 (core binding factor 1), a transcription factor playing a major role in the expression of osteoblastic specific genes, is involved in the expression of collagenase 3 during bone formation. We have functionally characterized a Cbfa motif present in the promoter region of collagenase 3 gene and demonstrated, by cotransfection experiments and gel mobility shift assays, that this element is involved in the inducibility of the collagenase 3 promoter by Cbfa1 in osteoblastic and chondrocytic cells. Furthermore, overexpression of Cbfa1 in osteoblastic cells unable to produce collagenase 3 leads to the expression of this gene after stimulation with transforming growth factor beta. Finally, we show that mutant mice deficient in Cbfa1, lacking mature osteoblasts but containing hypertrophic chondrocytes which are also a major source of collagenase 3, do not express this protease during fetal development. These results provide in vivo evidence that collagenase 3 is a target of the transcriptional activator Cbfa1 in these cells. On the basis of these transcriptional regulation studies, together with the potent proteolytic activity of collagenase 3 on diverse collagenous and noncollagenous bone and cartilage components, we proposed that this enzyme may play a key role in the process of bone formation and remodeling.
Figures


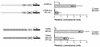



References
-
- Airola K, Johansson N, Kariniemi A L, Kähäri V M, Saarialho-Kere U K. Human collagenase-3 is expressed in malignant squamous epithelium of the skin. J Investig Dermatol. 1997;109:225–231. - PubMed
-
- Anglard P, Melot T, Guérin E, Thomas G, Basset P. Structure and promoter characterization of the human stromelysin-3 gene. J Biol Chem. 1995;270:20337–20344. - PubMed
-
- Balbín M, López-Otín C. Hormonal regulation of the human pepsinogen C gene in breast cancer cells: identification of a cis-acting element mediating its induction by androgens, glucocorticoids, and progesterone. J Biol Chem. 1996;271:15175–15181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
