Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: evidence for direct transcriptional repression of the Rb gene during differentiation
- PMID: 10330185
- PMCID: PMC104404
- DOI: 10.1128/MCB.19.6.4452
Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: evidence for direct transcriptional repression of the Rb gene during differentiation
Abstract
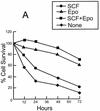
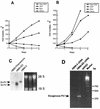
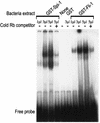
Erythropoietin (Epo) is a major regulator of erythropoiesis that alters the survival, proliferation, and differentiation of erythroid progenitor cells. The mechanism by which these events are regulated has not yet been determined. Using HB60, a newly established erythroblastic cell line, we show here that Epo-induced terminal erythroid differentiation is associated with a transient downregulation in the expression of the Ets-related transcription factor Fli-1. Constitutive expression of Fli-1 in HB60 cells, similar to retroviral insertional activation of Fli-1 observed in Friend murine leukemia virus (F-MuLV)-induced erythroleukemia, blocks Epo-induced differentiation while promoting Epo-induced proliferation. These results suggest that Fli-1 modulates the response of erythroid cells to Epo. To understand the mechanism by which Fli-1 regulates erythropoiesis, we searched for downstream target genes whose expression is regulated by this transcription factor. Here we show that the retinoblastoma (Rb) gene, which was previously shown to be involved in the development of mature erythrocytes, contains a Fli-1 consensus binding site within its promoter. Fli-1 binds to this cryptic Ets consensus site within the Rb promoter and transcriptionally represses Rb expression. Both the expression level and the phosphorylation status of Rb are consistent with the response of HB60 cells to Epo-induced terminal differentiation. We suggest that the negative regulation of Rb by Fli-1 could be one of the critical determinants in erythroid progenitor cell differentiation that is specifically deregulated during F-MuLV-induced erythroleukemia.
Figures



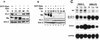
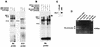
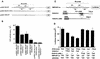
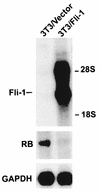

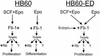
Similar articles
-
Epo regulates erythroid proliferation and differentiation through distinct signaling pathways: implication for erythropoiesis and Friend virus-induced erythroleukemia.Oncogene. 2000 May 4;19(19):2296-304. doi: 10.1038/sj.onc.1203590. Oncogene. 2000. PMID: 10822380
-
FLI-1 inhibits differentiation and induces proliferation of primary erythroblasts.Oncogene. 1999 Feb 25;18(8):1597-608. doi: 10.1038/sj.onc.1202534. Oncogene. 1999. PMID: 10102630
-
Up-regulation of SLAP in FLI-1-transformed erythroblasts interferes with EpoR signaling.Blood. 2003 Dec 15;102(13):4555-62. doi: 10.1182/blood-2003-06-2077. Epub 2003 Aug 28. Blood. 2003. PMID: 12946994
-
The role of Fli-1 in normal cell function and malignant transformation.Oncogene. 2000 Dec 18;19(55):6482-9. doi: 10.1038/sj.onc.1204042. Oncogene. 2000. PMID: 11175364 Review.
-
Role of c-Kit and erythropoietin receptor in erythropoiesis.Crit Rev Oncol Hematol. 2005 Apr;54(1):63-75. doi: 10.1016/j.critrevonc.2004.11.005. Crit Rev Oncol Hematol. 2005. PMID: 15780908 Review.
Cited by
-
An intricate regulatory circuit between FLI1 and GATA1/GATA2/LDB1/ERG dictates erythroid vs. megakaryocytic differentiation.Mol Med Rep. 2024 Jun;29(6):107. doi: 10.3892/mmr.2024.13231. Epub 2024 May 2. Mol Med Rep. 2024. PMID: 38695236 Free PMC article.
-
ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells.Blood. 2009 Apr 2;113(14):3337-47. doi: 10.1182/blood-2008-08-174813. Epub 2009 Jan 23. Blood. 2009. PMID: 19168790 Free PMC article.
-
Downregulation of Friend leukemia virus integration 1 as a feedback mechanism that restrains lipopolysaccharide induction of matrix metalloproteases and interleukin-10 in human macrophages.J Interferon Cytokine Res. 2010 Dec;30(12):893-900. doi: 10.1089/jir.2010.0046. Epub 2010 Sep 29. J Interferon Cytokine Res. 2010. PMID: 20879862 Free PMC article.
-
Overexpression of Fli-1 in astrocytoma is associated with poor prognosis.Oncotarget. 2017 Apr 25;8(17):29174-29186. doi: 10.18632/oncotarget.16303. Oncotarget. 2017. PMID: 28418872 Free PMC article.
-
An in silico proteomics screen to predict and prioritize protein-protein interactions dependent on post-translationally modified motifs.Bioinformatics. 2018 Nov 15;34(22):3898-3906. doi: 10.1093/bioinformatics/bty434. Bioinformatics. 2018. PMID: 29868839 Free PMC article.
References
-
- Ben-David Y, Giddens E G, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–918. - PubMed
-
- Ben-David Y, Prideaux V R, Chow V, Benchimol S, Bernstein A. Inactivation of the p53 oncogene by internal deletion or retroviral integration in erythroleukemic cell lines induced by Friend leukemia virus. Oncogene. 1988;3:179–185. - PubMed
-
- Bergh G, Ehinger M, Olofsson T, Baldetorp B, Johnsson E, Brycke H, Lindgren G, Olsson I, Gullberg U. Altered expression of the retinoblastoma tumor-suppressor gene in leukemic cell lines inhibits induction of differentiation but not G1-accumulation. Blood. 1997;89:2938–2950. - PubMed
-
- Chabot B, Stephenson D, Chapman V M, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335:88–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
