Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA
- PMID: 10339546
- PMCID: PMC26840
- DOI: 10.1073/pnas.96.11.6090
Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA
Abstract
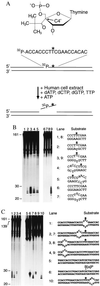
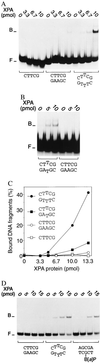
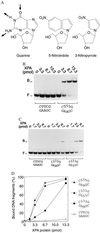
Nondistorting C4' backbone adducts serve as molecular tools to analyze the strategy by which a limited number of human nucleotide excision repair (NER) factors recognize an infinite variety of DNA lesions. We have constructed composite DNA substrates containing a noncomplementary site adjacent to a nondistorting C4' adduct to show that the loss of hydrogen bonding contacts between partner strands is an essential signal for the recruitment of NER enzymes. This specific conformational requirement for excision is mediated by the affinity of xeroderma pigmentosum group A (XPA) protein for nonhybridizing sites in duplex DNA. XPA recognizes defective Watson-Crick base pair conformations even in the absence of DNA adducts or other covalent modifications, apparently through detection of hydrophobic base components that are abnormally exposed to the double helical surface. This recognition function of XPA is enhanced by replication protein A (RPA) such that, in combination, XPA and RPA constitute a potent molecular sensor of denatured base pairs. Our results indicate that the XPA-RPA complex may promote damage recognition by monitoring Watson-Crick base pair integrity, thereby recruiting the human NER system preferentially to sites where hybridization between complementary strands is weakened or entirely disrupted.
Figures


References
-
- Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 634–649.
-
- Hoeijmakers J H J. Eur J Cancer. 1994;30:1912–1921. - PubMed
-
- Hanawalt P C. Science. 1994;266:1957–1958. - PubMed
-
- Sancar A. Annu Rev Biochem. 1996;65:43–81. - PubMed
-
- Wood R D. Annu Rev Biochem. 1996;65:135–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

