DNA methylation is a reversible biological signal
- PMID: 10339549
- PMCID: PMC26843
- DOI: 10.1073/pnas.96.11.6107
DNA methylation is a reversible biological signal
Abstract
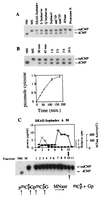
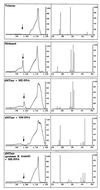
The pattern of DNA methylation plays an important role in regulating different genome functions. To test the hypothesis that DNA methylation is a reversible biochemical process, we purified a DNA demethylase from human cells that catalyzes the cleavage of a methyl residue from 5-methyl cytosine and its release as methanol. We show that similar to DNA methyltransferase, DNA demethylase shows CpG dinucleotide specificity, can demethylate mdCpdG sites in different sequence contexts, and demethylates both fully methylated and hemimethylated DNA. Thus, contrary to the commonly accepted model, DNA methylation is a reversible signal, similar to other physiological biochemical modifications.
Figures

Comment in
-
DNA demethylation.Proc Natl Acad Sci U S A. 1999 May 25;96(11):5894-6. doi: 10.1073/pnas.96.11.5894. Proc Natl Acad Sci U S A. 1999. PMID: 10339513 Free PMC article. Review. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

