Thermodynamic basis of the enhanced specificity of structured DNA probes
- PMID: 10339560
- PMCID: PMC26854
- DOI: 10.1073/pnas.96.11.6171
Thermodynamic basis of the enhanced specificity of structured DNA probes
Abstract
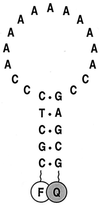
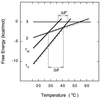
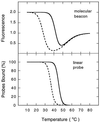
Molecular beacons are DNA probes that form a stem-and-loop structure and possess an internally quenched fluorophore. When they bind to complementary nucleic acids, they undergo a conformational transition that switches on their fluorescence. These probes recognize their targets with higher specificity than probes that cannot form a hairpin stem, and they easily discriminate targets that differ from one another by only a single nucleotide. Our results show that molecular beacons can exist in three different states: bound to a target, free in the form of a hairpin structure, and free in the form of a random coil. Thermodynamic analysis of the transitions between these states reveals that enhanced specificity is a general feature of conformationally constrained probes.
Figures



References
-
- Tyagi S, Kramer F R. Nat Biotechnol. 1996;14:303–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

