Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones
- PMID: 10339569
- PMCID: PMC26863
- DOI: 10.1073/pnas.96.11.6223
Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones
Abstract
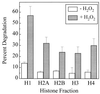
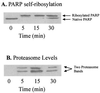
The 20S proteasome has been shown to be largely responsible for the degradation of oxidatively modified proteins in the cytoplasm. Nuclear proteins are also subject to oxidation, and the nucleus of mammalian cells contains proteasome. In human beings, tumor cells frequently are subjected to oxidation as a consequence of antitumor chemotherapy, and K562 human myelogenous leukemia cells have a higher nuclear proteasome activity than do nonmalignant cells. Adaptation to oxidative stress appears to be one element in the development of long-term resistance to many chemotherapeutic drugs and the mechanisms of inducible tumor resistance to oxidation are of obvious importance. After hydrogen peroxide treatment of K562 cells, degradation of the model proteasome peptide substrate suc-LLVY-MCA and degradation of oxidized histones in nuclei increases significantly within minutes. Both increased proteolytic susceptibility of the histone substrates (caused by modification by oxidation) and activation of the proteasome enzyme complex occur independently during oxidative stress. This rapid up-regulation of 20S proteasome activity is accompanied by, and depends on, poly-ADP ribosylation of the proteasome, as shown by inhibitor experiments, 14C-ADP ribose incorporation assays, immunoblotting, in vitro reconstitution experiments, and immunoprecipitation of (activated) proteasome with anti-poly-ADP ribose polymerase antibodies. The poly-ADP ribosylation-mediated activated nuclear 20S proteasome is able to remove oxidatively damaged histones more efficiently and therefore is proposed as an oxidant-stimulatable defense or repair system of the nucleus in K562 leukemia cells.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

