Direct genetic analysis by matrix-assisted laser desorption/ionization mass spectrometry
- PMID: 10339582
- PMCID: PMC26876
- DOI: 10.1073/pnas.96.11.6301
Direct genetic analysis by matrix-assisted laser desorption/ionization mass spectrometry
Abstract
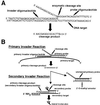
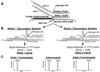
An approach to analyzing single-nucleotide polymorphisms (SNPs) found in the human genome has been developed that couples a recently developed invasive cleavage assay for nucleic acids with detection by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The invasive cleavage assay is a signal amplification method that enables the analysis of SNPs by MALDI-TOF MS directly from human genomic DNA without the need for initial target amplification by PCR. The results presented here show the successful genotyping by this approach of twelve SNPs located randomly throughout the human genome. Conventional Sanger sequencing of these SNP positions confirmed the accuracy of the MALDI-TOF MS analysis results. The ability to unambiguously detect both homozygous and heterozygous genotypes is clearly demonstrated. The elimination of the need for target amplification by PCR, combined with the inherently rapid and accurate nature of detection by MALDI-TOF MS, gives this approach unique and significant advantages in the high-throughput genotyping of large numbers of SNPs, useful for locating, identifying, and characterizing the function of specific genes.
Figures

References
-
- Wang D G, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1082. - PubMed
-
- Schafer A J, Hawkins J R. Nat Biotechnol. 1998;16:33–39. - PubMed
-
- Kruglyak L. Nat Genet. 1997;17:21–24. - PubMed
-
- Landegren U, Nilsson M, Kwok P-Y. Genome Res. 1998;8:769–776. - PubMed
-
- Karas M, Hillenkamp F. Anal Chem. 1988;60:2299–2303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

