Plasmodium falciparum subtilisin-like protease 2, a merozoite candidate for the merozoite surface protein 1-42 maturase
- PMID: 10339607
- PMCID: PMC26901
- DOI: 10.1073/pnas.96.11.6445
Plasmodium falciparum subtilisin-like protease 2, a merozoite candidate for the merozoite surface protein 1-42 maturase
Abstract
The process of human erythrocyte invasion by Plasmodium falciparum parasites involves a calcium-dependent serine protease with properties consistent with a subtilisin-like activity. This enzyme achieves the last crucial maturation step of merozoite surface protein 1 (MSP1) necessary for parasite entry into the host erythrocyte. In eukaryotic cells, such processing steps are performed by subtilisin-like maturases, known as proprotein convertases. In an attempt to characterize the MSP1 maturase, we have identified a gene that encodes a P. falciparum subtilisin-like protease (PfSUB2) whose deduced active site sequence resembles more bacterial subtilisins. Therefore, we propose that PfSUB2 belongs to a subclass of eukaryotic subtilisins different from proprotein convertases. Pfsub2 is expressed during merozoite differentiation and encodes an integral membrane protein localized in the merozoite dense granules, a secretory organelle whose contents are believed to participate in a late step of the erythrocyte invasion. PfSUB2's subcellular localization, together with its predicted enzymatic properties, leads us to propose that PfSUB2 could be responsible for the late MSP1 maturation step and thus is an attractive target for the development of new antimalarial drugs.
Figures

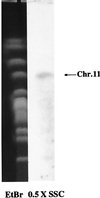

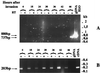

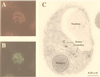

References
-
- Aikawa M. In: Malaria: Principles and Practice of Malariology. Wernsdorfer W H, McGregor I, editors. Vol. 1. Edinburgh: Churchill Livingstone; 1988. pp. 97–129.
-
- Bannister L H, Mitchell G H. J Protozool. 1989;36:362–367. - PubMed
-
- Braun-Breton C, Pereira da Silva L. Parasitol Today. 1993;9:92–96. - PubMed
-
- Perrin L H, Ramirez E, Lambert P H, Miescher P. Nature (London) 1981;289:301–303. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

