Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding
- PMID: 10346815
- PMCID: PMC316729
- DOI: 10.1101/gad.13.10.1263
Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding
Abstract
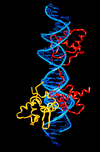
Pax6, a transcription factor containing the bipartite paired DNA-binding domain, has critical roles in development of the eye, nose, pancreas, and central nervous system. The 2.5 A structure of the human Pax6 paired domain with its optimal 26-bp site reveals extensive DNA contacts from the amino-terminal subdomain, the linker region, and the carboxy-terminal subdomain. The Pax6 structure not only confirms the docking arrangement of the amino-terminal subdomain as seen in cocrystals of the Drosophila Prd Pax protein, but also reveals some interesting differences in this region and helps explain the sequence specificity of paired domain-DNA recognition. In addition, this structure gives the first detailed information about how the paired linker region and carboxy-terminal subdomain contact DNA. The extended linker makes minor groove contacts over an 8-bp region, and the carboxy-terminal helix-turn-helix unit makes base contacts in the major groove. The structure and docking arrangement of the carboxy-terminal subdomain of Pax6 is remarkably similar to that of the amino-terminal subdomain, and there is an approximate twofold symmetry axis relating the polypeptide backbones of these two helix-turn-helix units. Our structure of the Pax6 paired domain-DNA complex provides a framework for understanding paired domain-DNA interactions, for analyzing mutations that map in the linker and carboxy-terminal regions of the paired domain, and for modeling protein-protein interactions of the Pax family proteins.
Figures


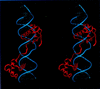


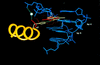
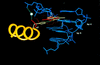
References
-
- Adams B, Dörfler P, Aguzzi A, Kozmik Z, Urbánek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes & Dev. 1992;6:1589–1607. - PubMed
-
- Azuma N, Nishina S, Yanagisawa H, Okuyama T, Yamada M. PAX6 missense mutation in isolated foveal hypoplasia. Nat Genet. 1996;18:141–142. - PubMed
-
- Azuma N, Hotta Y, Tanaka H, Yamada M. Missense mutations in the PAX6 gene in Aniridia. Invest Ophthalmol Vis Sci. 1998;39:2524–2528. - PubMed
-
- Baldwin CT, Hoth CF, Macina RA, Milunsky A. Mutations in Pax3 that cause Waardenburg syndrome type I: Ten new mutations and review of the literature. Am J Med Genet. 1995;58:115–122. - PubMed
-
- Barr FG. Chromosomal translocations involving paired box transcription factors in human cancer. Int J Biochem Cell Biol. 1997;29:1449–1461. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
