Assembly of functionally active Drosophila origin recognition complex from recombinant proteins
- PMID: 10346817
- PMCID: PMC316721
- DOI: 10.1101/gad.13.10.1289
Assembly of functionally active Drosophila origin recognition complex from recombinant proteins
Abstract
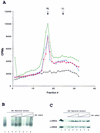
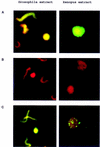
In eukaryotes the sites for the initiation of chromosomal DNA replication are believed to be determined in part by the binding of a heteromeric origin recognition complex (ORC) to DNA. We have cloned the genes encoding the subunits of the Drosophila ORC. Each of the genes is unique and can be mapped to discrete chromosomal locations implying that the pattern and developmental regulation of origin usage in Drosophila is not regulated solely by a large family of different ORC proteins. The six-subunit ORC can be reconstituted with recombinant proteins into a complex that restores DNA replication in ORC-depleted Drosophila or Xenopus egg extracts.
Figures



Similar articles
-
Interaction between the origin recognition complex and the replication licensing system in Xenopus.Cell. 1996 Oct 18;87(2):287-96. doi: 10.1016/s0092-8674(00)81346-x. Cell. 1996. PMID: 8861912
-
Role for a Xenopus Orc2-related protein in controlling DNA replication.Nature. 1996 Jan 25;379(6563):357-60. doi: 10.1038/379357a0. Nature. 1996. PMID: 8552193
-
Forced binding of the origin of replication complex to chromosomal sites in Drosophila S2 cells creates an origin of replication.J Cell Sci. 2012 Feb 15;125(Pt 4):965-72. doi: 10.1242/jcs.094409. Epub 2012 Mar 15. J Cell Sci. 2012. PMID: 22421364
-
Multiple functions of the origin recognition complex.Int Rev Cytol. 2007;256:69-109. doi: 10.1016/S0074-7696(07)56003-1. Int Rev Cytol. 2007. PMID: 17241905 Review.
-
ORC binding, gene amplification, and the nature of metazoan replication origins.Genes Dev. 1999 Oct 15;13(20):2619-23. doi: 10.1101/gad.13.20.2619. Genes Dev. 1999. PMID: 10541547 Review. No abstract available.
Cited by
-
Mechanisms and regulation of DNA replication initiation in eukaryotes.Crit Rev Biochem Mol Biol. 2017 Apr;52(2):107-144. doi: 10.1080/10409238.2016.1274717. Epub 2017 Jan 17. Crit Rev Biochem Mol Biol. 2017. PMID: 28094588 Free PMC article. Review.
-
Genetic interaction of an origin recognition complex subunit and the Polycomb group gene MEDEA during seed development.Plant Cell. 2004 Apr;16(4):1035-46. doi: 10.1105/tpc.019059. Epub 2004 Mar 12. Plant Cell. 2004. PMID: 15020747 Free PMC article.
-
Studies of the properties of human origin recognition complex and its Walker A motif mutants.Proc Natl Acad Sci U S A. 2005 Jan 4;102(1):69-74. doi: 10.1073/pnas.0408690102. Epub 2004 Dec 23. Proc Natl Acad Sci U S A. 2005. PMID: 15618391 Free PMC article.
-
DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding.EMBO J. 2004 Feb 25;23(4):897-907. doi: 10.1038/sj.emboj.7600077. Epub 2004 Feb 5. EMBO J. 2004. PMID: 14765124 Free PMC article.
-
The origin recognition complex is dispensable for endoreplication in Drosophila.Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12343-8. doi: 10.1073/pnas.0805189105. Epub 2008 Aug 18. Proc Natl Acad Sci U S A. 2008. PMID: 18711130 Free PMC article.
References
-
- Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. - PubMed
-
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. - PubMed
-
- ————— A role for the nuclear envelope in controling DNA replication within the cell cycle. Nature. 1988;332:546–548. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
