Polarity determination in the Drosophila eye: a novel role for unpaired and JAK/STAT signaling
- PMID: 10346822
- PMCID: PMC316719
- DOI: 10.1101/gad.13.10.1342
Polarity determination in the Drosophila eye: a novel role for unpaired and JAK/STAT signaling
Abstract
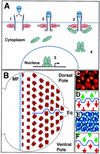
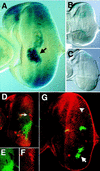
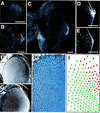
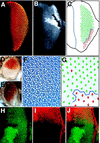
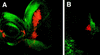
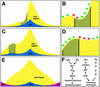
The JAK/STAT signaling pathway is required for many processes including cytokine signaling, hematopoiesis, gliagenesis, and Drosophila segmentation. In this report we present evidence demonstrating that the JAK/STAT pathway is also central to the establishment of planar polarity during Drosophila eye development. We show that a localized source of the pathway ligand, Unpaired, is present at the midline of the developing eye, which is capable of activating the JAK/STAT pathway over long distances. A gradient of JAK/STAT activity across the DV axis of the eye regulates ommatidial polarity via an unidentified second signal. Additionally, localized Unpaired influences the position of the equator via repression of mirror.
Figures

References
-
- Baker NE, Rubin GM. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev Biol. 1992;150:381–396. - PubMed
-
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes & Dev. 1994;8:300–312. - PubMed
-
- Bonni A, Sun Y, Nadal VM, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliagenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Brodsky MH, Steller H. Positional information along the dorsal-ventral axis of the Drosophila eye: Graded expression of the four-jointed gene. Dev Biol. 1996;173:428–446. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
