Chitinases from uncultured marine microorganisms
- PMID: 10347042
- PMCID: PMC91377
- DOI: 10.1128/AEM.65.6.2553-2557.1999
Chitinases from uncultured marine microorganisms
Abstract
Our understanding of the degradation of organic matter will benefit from a greater appreciation for the genes encoding enzymes involved in the hydrolysis of biopolymers such as chitin, one of the most abundant polymers in nature. To isolate representative and abundant chitinase genes from uncultivated marine bacteria, we constructed libraries of genomic DNA isolated from coastal and estuarine waters. The libraries were screened for genes encoding proteins that hydrolyze a fluorogenic analogue of chitin, 4-methylumbelliferyl beta-D-N,N'-diacetylchitobioside (MUF-diNAG). The abundance of clones capable of MUF-diNAG hydrolysis was higher in the library constructed with DNA from the estuary than in that constructed with DNA from coastal waters, although the abundance of positive clones was also dependent on the method used to screen the library. Plaque assays revealed nine MUF-diNAG-positive clones of 75,000 screened for the estuarine sample and two clones of 750,000 for the coastal sample. A microtiter plate assay revealed approximately 1 positive clone for every 500 clones screened in the coastal library. The number of clones detected with the plaque assay was consistent with estimates of the portion of culturable bacteria that degrade chitin. Our results suggest that culture-dependent methods do not greatly underestimate the portion of marine bacterial communities capable of chitin degradation.
Figures
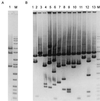
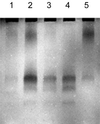
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struh K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1992.
-
- Bassler B L, Yu C, Lee Y C, Roseman S. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24276–24286. - PubMed
-
- Blackwell J, Parker K D, Rudall K M. Chitin fibers of the diatoms Thalassiosira fluviatilis and Cyclotella cryptica. J Mol Biol. 1967;28:383. - PubMed
-
- Brisou J, Tysset C, de Rautlin A, de la Roy Y, Curcier R, Moreau R. Etude sur la chitinolyse a milieu marin. Ann Inst Pasteur. 1964;106:469–478.
-
- Chretiennot-Dinet M J, Giraud-Guille M M, Vaulot D, Putaux J L, Saito Y, Chanzy H. The chitinous nature of filaments ejected by Phaeocystis (Prymnesiophyceae) J Phycol. 1997;33:666–672.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

