Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean
- PMID: 10347047
- PMCID: PMC91382
- DOI: 10.1128/AEM.65.6.2585-2591.1999
Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean
Abstract
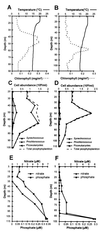
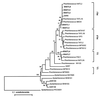
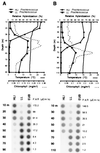
The in situ community structure of Prochlorococcus populations in the eastern North Atlantic Ocean was examined by analysis of Prochlorococcus 16S rDNA sequences with three independent approaches: cloning and sequencing, hybridization to specific oligonucleotide probes, and denaturing gradient gel electrophoresis (DGGE). The hybridization of high-light (HL) and low-light (LL) Prochlorococcus genotype-specific probes to two depth profiles of PCR-amplified 16S rDNA sequences revealed that in these two stratified water columns, an obvious niche-partitioning of Prochlorococcus genotypes occurred. In each water column a shift from the HL to the LL genotype was observed, a transition correlating with the depth of the surface mixed layer (SML). Only the HL genotype was found in the SML in each water column, whereas the LL genotype was distributed below the SML. The range of in situ irradiance to which each genotype was subjected within these distinct niches was consistent with growth irradiance studies of cultured HL- and LL-adapted Prochlorococcus strains. DGGE analysis and the sequencing of Prochlorococcus 16S rDNA clones were in full agreement with the genotype-specific oligonucleotide probe hybridization data. These observations of a partitioning of Prochlorococcus genotypes in a stratified water column provide a genetic basis for the dim and bright Prochlorococcus populations observed in flow cytometric signatures in several oceanic provinces.
Figures
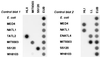

References
-
- Binder B J, Chisholm S W, Olson R J, Frankel S L, Worden A Z. Dynamics of picophytoplankton, ultraphytoplankton and bacteria in the central equatorial Pacific. Deep-Sea Res. 1996;43:907–931.
-
- Campbell L, Nolla H A, Vaulot D. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol Oceanogr. 1994;39:954–961.
-
- Campbell L, Vaulot D. Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (Station ALOHA) Deep-Sea Res. 1993;40:2043–2060.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

