Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli
- PMID: 10348852
- PMCID: PMC93807
- DOI: 10.1128/JB.181.11.3409-3418.1999
Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli
Abstract
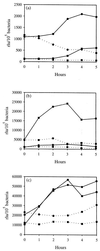
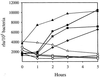
We have determined that the genes encoding the secreted proteins EspA, EspD, and EspB of enterohemorrhagic Escherichia coli (EHEC) are organized in a single operon. The esp operon is controlled by a promoter located 94 bp upstream from the ATG start codon of the espA gene. The promoter is activated in the early logarithmic growth phase, upon bacterial contact with eukaryotic cells and in response to Ca2+, Mn2+, and HEPES. Transcription of the esp operon seems to be switched off in tightly attached bacteria. The activation process is regulated by osmolarity (induction at high osmolarities), modulated by temperature, and influenced by the degree of DNA supercoiling. Transcription is sigmaS dependent, and the H-NS protein contributes to its fine tuning. Identification of the factors involved in activation of the esp operon and the signals responsible for modulation may facilitate understanding of the underlying molecular events leading to sequential expression of virulence factors during natural infections caused by EHEC.
Figures






Similar articles
-
Characterization of SepL of enterohemorrhagic Escherichia coli.J Bacteriol. 2000 Nov;182(22):6490-8. doi: 10.1128/JB.182.22.6490-6498.2000. J Bacteriol. 2000. PMID: 11053395 Free PMC article.
-
Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7.J Bacteriol. 2004 Nov;186(21):7290-301. doi: 10.1128/JB.186.21.7290-7301.2004. J Bacteriol. 2004. PMID: 15489441 Free PMC article.
-
Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level.Infect Immun. 2003 Oct;71(10):5900-9. doi: 10.1128/IAI.71.10.5900-5909.2003. Infect Immun. 2003. PMID: 14500511 Free PMC article.
-
Regulation of Escherichia coli Pathogenesis by Alternative Sigma Factor N.EcoSal Plus. 2017 Jun;7(2):10.1128/ecosalplus.ESP-0016-2016. doi: 10.1128/ecosalplus.ESP-0016-2016. EcoSal Plus. 2017. PMID: 28635589 Free PMC article. Review.
-
Advances in bacterial transcriptome understanding: From overlapping transcription to the excludon concept.Mol Microbiol. 2020 Mar;113(3):593-602. doi: 10.1111/mmi.14456. Mol Microbiol. 2020. PMID: 32185833 Free PMC article. Review.
Cited by
-
H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues.J Bacteriol. 2000 Nov;182(22):6391-400. doi: 10.1128/JB.182.22.6391-6400.2000. J Bacteriol. 2000. PMID: 11053383 Free PMC article.
-
Optimization of reverse transcriptase PCR to detect viable Shiga-toxin-producing Escherichia coli.Appl Environ Microbiol. 2002 Feb;68(2):799-806. doi: 10.1128/AEM.68.2.799-806.2002. Appl Environ Microbiol. 2002. PMID: 11823221 Free PMC article.
-
Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12.J Bacteriol. 2009 Feb;191(3):1106-10. doi: 10.1128/JB.00599-08. Epub 2008 Nov 14. J Bacteriol. 2009. PMID: 19011022 Free PMC article.
-
Characterization of SepL of enterohemorrhagic Escherichia coli.J Bacteriol. 2000 Nov;182(22):6490-8. doi: 10.1128/JB.182.22.6490-6498.2000. J Bacteriol. 2000. PMID: 11053395 Free PMC article.
-
DNA Supercoiling: an Ancestral Regulator of Gene Expression in Pathogenic Bacteria?Comput Struct Biotechnol J. 2019 Jul 26;17:1047-1055. doi: 10.1016/j.csbj.2019.07.013. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31452857 Free PMC article. Review.
References
-
- Akerley J F, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. - PubMed
-
- Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. - PubMed
-
- Boyce T G, Swerdlow D L, Griffin P M. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N Engl J Med. 1995;333:364–368. - PubMed
-
- Conter A, Menchon C, Gutierrez C. Role of DNA supercoiling and RpoS sigma factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J Mol Biol. 1997;273:75–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

