Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae
- PMID: 10348861
- PMCID: PMC93816
- DOI: 10.1128/JB.181.11.3478-3485.1999
Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae
Abstract
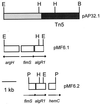
Both Pseudomonas aeruginosa and the phytopathogen P. syringae produce the exopolysaccharide alginate. However, the environmental signals that trigger alginate gene expression in P. syringae are different from those in P. aeruginosa with copper being a major signal in P. syringae. In P. aeruginosa, the alternate sigma factor encoded by algT (sigma22) and the response regulator AlgR1 are required for transcription of algD, a gene which encodes a key enzyme in the alginate biosynthetic pathway. In the present study, we cloned and characterized the gene encoding AlgR1 from P. syringae. The deduced amino acid sequence of AlgR1 from P. syringae showed 86% identity to its P. aeruginosa counterpart. Sequence analysis of the region flanking algR1 in P. syringae revealed the presence of argH, algZ, and hemC in an arrangement virtually identical to that reported in P. aeruginosa. An algR1 mutant, P. syringae FF5.32, was defective in alginate production but could be complemented when algR1 was expressed in trans. The algD promoter region in P. syringae (PsalgD) was also characterized and shown to diverge significantly from the algD promoter in P. aeruginosa. Unlike P. aeruginosa, algR1 was not required for the transcription of algD in P. syringae, and PsalgD lacked the consensus sequence recognized by AlgR1. However, both the algD and algR1 upstream regions in P. syringae contained the consensus sequence recognized by sigma22, suggesting that algT is required for transcription of both genes.
Figures

Similar articles
-
AlgT (sigma22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae.J Bacteriol. 1999 Dec;181(23):7176-84. doi: 10.1128/JB.181.23.7176-7184.1999. J Bacteriol. 1999. PMID: 10572118 Free PMC article.
-
AlgR functions in algC expression and virulence in Pseudomonas syringae pv. syringae.Microbiology (Reading). 2004 Aug;150(Pt 8):2727-2737. doi: 10.1099/mic.0.27199-0. Microbiology (Reading). 2004. PMID: 15289569
-
Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae.J Bacteriol. 1997 Jul;179(14):4464-72. doi: 10.1128/jb.179.14.4464-4472.1997. J Bacteriol. 1997. PMID: 9226254 Free PMC article.
-
Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT.J Bacteriol. 1994 Oct;176(19):6007-14. doi: 10.1128/jb.176.19.6007-6014.1994. J Bacteriol. 1994. PMID: 7928961 Free PMC article.
-
The algD promoter: regulation of alginate production by Pseudomonas aeruginosa in cystic fibrosis.Cell Mol Biol Res. 1993;39(4):371-6. Cell Mol Biol Res. 1993. PMID: 8312973 Review.
Cited by
-
The algT gene of Pseudomonas syringae pv. glycinea and new insights into the transcriptional organization of the algT-muc gene cluster.J Bacteriol. 2006 Dec;188(23):8013-21. doi: 10.1128/JB.01160-06. Epub 2006 Sep 29. J Bacteriol. 2006. PMID: 17012388 Free PMC article.
-
Characterization of the transcriptional activators SalA and SyrF, Which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae.J Bacteriol. 2006 May;188(9):3290-8. doi: 10.1128/JB.188.9.3290-3298.2006. J Bacteriol. 2006. PMID: 16621822 Free PMC article.
-
Biological role of EPS from Pseudomonas syringae pv. syringae UMAF0158 extracellular matrix, focusing on a Psl-like polysaccharide.NPJ Biofilms Microbiomes. 2020 Oct 12;6(1):37. doi: 10.1038/s41522-020-00148-6. NPJ Biofilms Microbiomes. 2020. PMID: 33046713 Free PMC article.
-
Pseudomonas biofilm matrix composition and niche biology.FEMS Microbiol Rev. 2012 Jul;36(4):893-916. doi: 10.1111/j.1574-6976.2011.00322.x. Epub 2012 Jan 23. FEMS Microbiol Rev. 2012. PMID: 22212072 Free PMC article. Review.
-
AlgT (sigma22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae.J Bacteriol. 1999 Dec;181(23):7176-84. doi: 10.1128/JB.181.23.7176-7184.1999. J Bacteriol. 1999. PMID: 10572118 Free PMC article.
References
-
- Aarons S J, Sutherland I W, Chakrabarty A M, Gallagher M P. A novel gene, algK, from the alginate biosynthetic cluster of Pseudomonas aeruginosa. Microbiology. 1997;143:641–652. - PubMed
-
- Baynham P J, Wozniak D J. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

