Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter
- PMID: 10348863
- PMCID: PMC93818
- DOI: 10.1128/JB.181.11.3494-3504.1999
Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter
Abstract
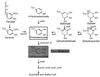
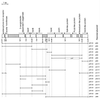
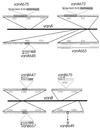
VanA and VanB form an oxygenative demethylase that converts vanillate to protocatechuate in microorganisms. Ferulate, an abundant phytochemical, had been shown to be metabolized through a vanillate intermediate in several Pseudomonas isolates, and biochemical evidence had indicated that vanillate also is an intermediate in ferulate catabolism by Acinetobacter. Genetic evidence supporting this conclusion was obtained by characterization of mutant Acinetobacter strains blocked in catabolism of both ferulate and vanillate. Cloned Acinetobacter vanA and vanB were shown to be members of a chromosomal segment remote from a supraoperonic cluster containing other genes required for completion of the catabolism of ferulate and its structural analogs, caffeate and coumarate, through protocatechuate. The nucleotide sequence of DNA containing vanA and vanB demonstrated the presence of genes that, on the basis of nucleotide sequence similarity, appeared to be associated with transport of aromatic compounds, metabolism of such compounds, or iron scavenging. Spontaneous deletion of 100 kb of DNA containing this segment does not impede the growth of cells with simple carbon sources other than vanillate or ferulate. Additional spontaneous mutations blocking vanA and vanB expression were shown to be mediated by IS1236, including insertion of the newly discovered composite transposon Tn5613. On the whole, vanA and vanB appear to be located within a nonessential genetic region that exhibits considerable genetic malleability in Acinetobacter. The overall organization of genes neighboring Acinetobacter vanA and vanB, including a putative transcriptional regulatory gene that is convergently transcribed and overlaps vanB, is conserved in Pseudomonas aeruginosa but has undergone radical rearrangement in other Pseudomonas species.
Figures


Similar articles
-
Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate.J Bacteriol. 1997 Apr;179(8):2595-607. doi: 10.1128/jb.179.8.2595-2607.1997. J Bacteriol. 1997. PMID: 9098058 Free PMC article.
-
Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13.Appl Environ Microbiol. 2000 Jun;66(6):2311-7. doi: 10.1128/AEM.66.6.2311-2317.2000. Appl Environ Microbiol. 2000. PMID: 10831404 Free PMC article.
-
Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase.J Bacteriol. 1988 Oct;170(10):4924-30. doi: 10.1128/jb.170.10.4924-4930.1988. J Bacteriol. 1988. PMID: 3170489 Free PMC article.
-
The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK.J Bacteriol. 1999 Jun;181(11):3505-15. doi: 10.1128/JB.181.11.3505-3515.1999. J Bacteriol. 1999. PMID: 10348864 Free PMC article.
-
Substrate range and genetic analysis of Acinetobacter vanillate demethylase.J Bacteriol. 2000 Mar;182(5):1383-9. doi: 10.1128/JB.182.5.1383-1389.2000. J Bacteriol. 2000. PMID: 10671462 Free PMC article.
Cited by
-
Metabolism of isovanillate, vanillate, and veratrate by Comamonas testosteroni strain BR6020.J Bacteriol. 2006 Jun;188(11):3862-9. doi: 10.1128/JB.01675-05. J Bacteriol. 2006. PMID: 16707678 Free PMC article.
-
A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus.Nucleic Acids Res. 2007;35(20):e137. doi: 10.1093/nar/gkm818. Epub 2007 Oct 24. Nucleic Acids Res. 2007. PMID: 17959646 Free PMC article.
-
Genes for chlorogenate and hydroxycinnamate catabolism (hca) are linked to functionally related genes in the dca-pca-qui-pob-hca chromosomal cluster of Acinetobacter sp. strain ADP1.Appl Environ Microbiol. 2003 Jan;69(1):524-32. doi: 10.1128/AEM.69.1.524-532.2003. Appl Environ Microbiol. 2003. PMID: 12514037 Free PMC article.
-
Involvement of a PadR regulator PrhP on virulence of Ralstonia solanacearum by controlling detoxification of phenolic acids and type III secretion system.Mol Plant Pathol. 2019 Nov;20(11):1477-1490. doi: 10.1111/mpp.12854. Epub 2019 Aug 8. Mol Plant Pathol. 2019. PMID: 31392803 Free PMC article.
-
Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis.Biomolecules. 2020 Jun 6;10(6):874. doi: 10.3390/biom10060874. Biomolecules. 2020. PMID: 32517243 Free PMC article. Review.
References
-
- Bouvet P J, Jeanjean S. Differentiation of Acinetobacter calcoaceticus sensu stricto from related Acinetobacter species by electrophoretic polymorphism of malate dehydrogenase, glutamate dehydrogenase and catalase. Res Microbiol. 1995;146:773–785. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

