The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK
- PMID: 10348864
- PMCID: PMC93819
- DOI: 10.1128/JB.181.11.3505-3515.1999
The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK
Abstract
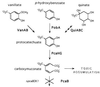
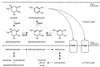
VanK is the fourth member of the ubiquitous major facilitator superfamily of transport proteins to be identified that, together with PcaK, BenK, and MucK, contributes to aromatic catabolism in Acinetobacter sp. strain ADP1. VanK and PcaK have overlapping specificity for p-hydroxybenzoate and, most clearly, for protocatechuate: inactivation of both proteins severely impairs growth with protocatechuate, and the activity of either protein alone can mask the phenotype associated with inactivation of its homolog. Furthermore, vanK pcaK double-knockout mutants appear completely unable to grow in liquid culture with the hydroaromatic compound quinate, although such cells on plates convert quinate to protocatechuate, which then accumulates extracellularly and is readily visible as purple staining. This provides genetic evidence that quinate is converted to protocatechuate in the periplasm and is in line with the early argument that quinate catabolism should be physically separated from aromatic amino acid biosynthesis in the cytoplasm so as to avoid potential competition for intermediates common to both pathways. Previous studies of aromatic catabolism in Acinetobacter have taken advantage of the ability to select directly strains that contain a spontaneous mutation blocking the beta-ketoadipate pathway and preventing the toxic accumulation of carboxymuconate. By using this procedure, strains with a mutation in structural or regulatory genes blocking degradation of vanillate, p-hydroxybenzoate, or protocatechuate were selected. In this study, the overlapping specificity of the VanK and PcaK permeases was exploited to directly select strains with a mutation in either vanK or pcaK. Spontaneous mutations identified in vanK include a hot spot for frameshift mutation due to contraction of a G6 mononucleotide repeat as well as point mutations producing amino acid substitutions useful for analysis of VanK structure and function. Preliminary second-site suppression analysis using transformation-facilitated PCR mutagenesis in one VanK mutant gave results similar to those using LacY, the prototypic member of the major facilitator superfamily, consistent with the two proteins having a similar mechanism of action. The selection for transport mutants described here for Acinetobacter may also be applicable to Pseudomonas putida, where the PcaK permease has an additional role in chemotaxis.
Figures





Similar articles
-
PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter.J Bacteriol. 1998 Mar;180(6):1512-24. doi: 10.1128/JB.180.6.1512-1524.1998. J Bacteriol. 1998. PMID: 9515921 Free PMC article.
-
PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida.J Bacteriol. 1997 Aug;179(16):5056-61. doi: 10.1128/jb.179.16.5056-5061.1997. J Bacteriol. 1997. PMID: 9260946 Free PMC article.
-
Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum.Microbiology (Reading). 2007 Mar;153(Pt 3):857-865. doi: 10.1099/mic.0.2006/002501-0. Microbiology (Reading). 2007. PMID: 17322206
-
Biophysical analyses of designed and selected mutants of protocatechuate 3,4-dioxygenase1.Annu Rev Microbiol. 2004;58:555-85. doi: 10.1146/annurev.micro.57.030502.090927. Annu Rev Microbiol. 2004. PMID: 15487948 Review.
-
From membrane to molecule to the third amino acid from the left with a membrane transport protein.Q Rev Biophys. 1997 Nov;30(4):333-64. doi: 10.1017/s0033583597003387. Q Rev Biophys. 1997. PMID: 9634651 Review.
Cited by
-
Transcriptional cross-regulation of the catechol and protocatechuate branches of the beta-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. strain ADP1 pobA gene.Appl Environ Microbiol. 2003 Mar;69(3):1598-606. doi: 10.1128/AEM.69.3.1598-1606.2003. Appl Environ Microbiol. 2003. PMID: 12620848 Free PMC article.
-
Cloning and genetic characterization of dca genes required for beta-oxidation of straight-chain dicarboxylic acids in Acinetobacter sp. strain ADP1.Appl Environ Microbiol. 2001 Oct;67(10):4817-27. doi: 10.1128/AEM.67.10.4817-4827.2001. Appl Environ Microbiol. 2001. PMID: 11571189 Free PMC article.
-
Characterization of Highly Ferulate-Tolerant Acinetobacter baylyi ADP1 Isolates by a Rapid Reverse Engineering Method.Appl Environ Microbiol. 2022 Jan 25;88(2):e0178021. doi: 10.1128/AEM.01780-21. Epub 2021 Nov 17. Appl Environ Microbiol. 2022. PMID: 34788063 Free PMC article.
-
Transcriptional organization of genes for protocatechuate and quinate degradation from Acinetobacter sp. strain ADP1.Appl Environ Microbiol. 2005 Feb;71(2):1025-34. doi: 10.1128/AEM.71.2.1025-1034.2005. Appl Environ Microbiol. 2005. PMID: 15691962 Free PMC article.
-
A second 5-carboxyvanillate decarboxylase gene, ligW2, is important for lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6.Appl Environ Microbiol. 2005 Sep;71(9):5014-21. doi: 10.1128/AEM.71.9.5014-5021.2005. Appl Environ Microbiol. 2005. PMID: 16151081 Free PMC article.
References
-
- Bray D, Levin M D, Morton-Firth C J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

