Control of acid resistance in Escherichia coli
- PMID: 10348866
- PMCID: PMC93821
- DOI: 10.1128/JB.181.11.3525-3535.1999
Control of acid resistance in Escherichia coli
Abstract
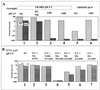
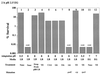
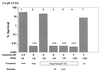
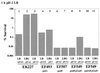
Acid resistance (AR) in Escherichia coli is defined as the ability to withstand an acid challenge of pH 2.5 or less and is a trait generally restricted to stationary-phase cells. Earlier reports described three AR systems in E. coli. In the present study, the genetics and control of these three systems have been more clearly defined. Expression of the first AR system (designated the oxidative or glucose-repressed AR system) was previously shown to require the alternative sigma factor RpoS. Consistent with glucose repression, this system also proved to be dependent in many situations on the cyclic AMP receptor protein. The second AR system required the addition of arginine during pH 2.5 acid challenge, the structural gene for arginine decarboxylase (adiA), and the regulator cysB, confirming earlier reports. The third AR system required glutamate for protection at pH 2.5, one of two genes encoding glutamate decarboxylase (gadA or gadB), and the gene encoding the putative glutamate:gamma-aminobutyric acid antiporter (gadC). Only one of the two glutamate decarboxylases was needed for protection at pH 2.5. However, survival at pH 2 required both glutamate decarboxylase isozymes. Stationary phase and acid pH regulation of the gad genes proved separable. Stationary-phase induction of gadA and gadB required the alternative sigma factor sigmaS encoded by rpoS. However, acid induction of these enzymes, which was demonstrated to occur in exponential- and stationary-phase cells, proved to be sigmaS independent. Neither gad gene required the presence of volatile fatty acids for induction. The data also indicate that AR via the amino acid decarboxylase systems requires more than an inducible decarboxylase and antiporter. Another surprising finding was that the sigmaS-dependent oxidative system, originally thought to be acid induced, actually proved to be induced following entry into stationary phase regardless of the pH. However, an inhibitor produced at pH 8 somehow interferes with the activity of this system, giving the illusion of acid induction. The results also revealed that the AR system affording the most effective protection at pH 2 in complex medium (either Luria-Bertani broth or brain heart infusion broth plus 0.4% glucose) is the glutamate-dependent GAD system. Thus, E. coli possesses three overlapping acid survival systems whose various levels of control and differing requirements for activity ensure that at least one system will be available to protect the stationary-phase cell under naturally occurring acidic environments.
Figures



References
-
- De Lorenzo V C, Timmis K M. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

