Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis
- PMID: 10357856
- PMCID: PMC98971
- DOI: 10.1128/MMBR.63.2.405-445.1999
Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis
Abstract
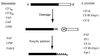
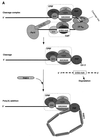
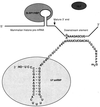
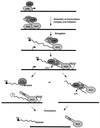
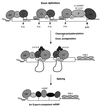
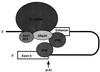
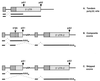
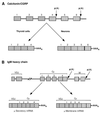
Formation of mRNA 3' ends in eukaryotes requires the interaction of transacting factors with cis-acting signal elements on the RNA precursor by two distinct mechanisms, one for the cleavage of most replication-dependent histone transcripts and the other for cleavage and polyadenylation of the majority of eukaryotic mRNAs. Most of the basic factors have now been identified, as well as some of the key protein-protein and RNA-protein interactions. This processing can be regulated by changing the levels or activity of basic factors or by using activators and repressors, many of which are components of the splicing machinery. These regulatory mechanisms act during differentiation, progression through the cell cycle, or viral infections. Recent findings suggest that the association of cleavage/polyadenylation factors with the transcriptional complex via the carboxyl-terminal domain of the RNA polymerase II (Pol II) large subunit is the means by which the cell restricts polyadenylation to Pol II transcripts. The processing of 3' ends is also important for transcription termination downstream of cleavage sites and for assembly of an export-competent mRNA. The progress of the last few years points to a remarkable coordination and cooperativity in the steps leading to the appearance of translatable mRNA in the cytoplasm.
Figures




References
-
- Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

