Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris
- PMID: 10359594
- PMCID: PMC25367
- DOI: 10.1091/mbc.10.6.1745
Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris
Abstract
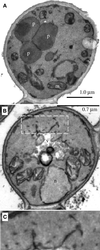
We report the cloning and characterization of Pichia pastoris PEX19 by complementation of a peroxisome-deficient mutant strain. Import of peroxisomal targeting signal 1- and 2-containing peroxisomal matrix proteins is defective in pex19 mutants. PEX19 encodes a hydrophilic 299-amino acid protein with sequence similarity to Saccharomyces cerevisiae Pex19p and human and Chinese hamster PxF, all farnesylated proteins, as well as hypothetical proteins from Caenorhabditis elegans and Schizosaccharomyces pombe. The farnesylation consensus is conserved in PpPex19p but dispensable for function and appears unmodified under the conditions tested. Pex19p localizes predominantly to the cytosolic fraction. Biochemical and two-hybrid analyses confirmed that Pex19p interacts with Pex3p, as seen in S. cerevisiae, but unexpectedly also with Pex10p. Two-hybrid analysis demonstrated that the amino-terminal 42 amino acids of Pex19p interact with the carboxyl-terminal 335 amino acids of Pex3p. In addition, the extreme carboxyl terminus of Pex19p (67 amino acids) is required for interaction with the amino-terminal 380 amino acids of Pex10p. Biochemical and immunofluorescence microscopy analyses of pex19Delta cells identified the membrane protein Pex3p in peroxisome remnants that were not previously observed in S. cerevisiae. These small vesicular and tubular (early) remnants are morphologically distinct from other Pppex mutant (late) remnants, suggesting that Pex19p functions at an early stage of peroxisome biogenesis.
Figures











Similar articles
-
Pex17p is required for import of both peroxisome membrane and lumenal proteins and interacts with Pex19p and the peroxisome targeting signal-receptor docking complex in Pichia pastoris.Mol Biol Cell. 1999 Dec;10(12):4005-19. doi: 10.1091/mbc.10.12.4005. Mol Biol Cell. 1999. PMID: 10588639 Free PMC article.
-
A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane.J Biol Chem. 2000 Apr 7;275(14):9986-95. doi: 10.1074/jbc.275.14.9986. J Biol Chem. 2000. PMID: 10744674
-
Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris.J Biol Chem. 1996 Aug 2;271(31):18973-80. doi: 10.1074/jbc.271.31.18973. J Biol Chem. 1996. PMID: 8702562
-
Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease.Biochim Biophys Acta. 2012 Sep;1822(9):1337-42. doi: 10.1016/j.bbadis.2012.06.004. Epub 2012 Jun 13. Biochim Biophys Acta. 2012. PMID: 22705440 Review.
-
Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions.Biochim Biophys Acta. 2006 Dec;1763(12):1639-46. doi: 10.1016/j.bbamcr.2006.09.030. Epub 2006 Sep 26. Biochim Biophys Acta. 2006. PMID: 17069900 Review.
Cited by
-
The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane.J Cell Biol. 2000 Jun 12;149(6):1171-8. doi: 10.1083/jcb.149.6.1171. J Cell Biol. 2000. PMID: 10851015 Free PMC article.
-
Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway.J Cell Biol. 2013 Mar 4;200(5):651-66. doi: 10.1083/jcb.201211077. J Cell Biol. 2013. PMID: 23460677 Free PMC article.
-
Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner.Mol Biol Cell. 2000 Nov;11(11):3963-76. doi: 10.1091/mbc.11.11.3963. Mol Biol Cell. 2000. PMID: 11071920 Free PMC article.
-
Farnesylation of pex19p is required for its structural integrity and function in peroxisome biogenesis.J Biol Chem. 2009 Jul 31;284(31):20885-96. doi: 10.1074/jbc.M109.016584. Epub 2009 May 18. J Biol Chem. 2009. PMID: 19451657 Free PMC article.
-
Peroxisome assembly: matrix and membrane protein biogenesis.J Cell Biol. 2011 Apr 4;193(1):7-16. doi: 10.1083/jcb.201010022. J Cell Biol. 2011. PMID: 21464226 Free PMC article. Review.
References
-
- Baerends RJS, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PTW, Kiel JAKW, Cregg JM, van der Klei I, Veenhuis M. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. - PubMed
-
- Braun A, Kammerer S, Weissenhorn W, Weiss EH, Cleve H. Sequence of a putative human housekeeping gene (HK33) localized on chromosome 1. Gene. 1994;146:291–295. - PubMed
-
- De Duve C. The peroxisome in retrospect. Ann NY Acad Sci. 1996;804:1–10. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

