Deciphering the nuclear import pathway for the cytoskeletal red cell protein 4.1R
- PMID: 10359596
- PMCID: PMC25371
- DOI: 10.1091/mbc.10.6.1783
Deciphering the nuclear import pathway for the cytoskeletal red cell protein 4.1R
Abstract
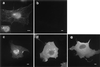
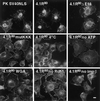
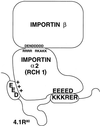
The erythroid membrane cytoskeletal protein 4.1 is the prototypical member of a genetically and topologically complex family that is generated by combinatorial alternative splicing pathways and is localized at diverse intracellular sites including the nucleus. To explore the molecular determinants for nuclear localization, we transfected COS-7 cells with epitope-tagged versions of natural red cell protein 4.1 (4.1R) isoforms as well as mutagenized and truncated derivatives. Two distant topological sorting signals were required for efficient nuclear import of the 4.1R80 isoform: a basic peptide, KKKRER, encoded by alternative exon 16 and acting as a weak core nuclear localization signal (4.1R NLS), and an acidic peptide, EED, encoded by alternative exon 5. 4.1R80 isoforms lacking either of these two exons showed decreased nuclear import. Fusion of various 4.1R80 constructs to the cytoplasmic reporter protein pyruvate kinase confirmed a requirement for both motifs for full NLS function. 4.1R80 was efficiently imported in the nuclei of digitonin-permeabilized COS-7 cells in the presence of recombinant Rch1 (human importin alpha2), importin beta, and GTPase Ran. Quantitative analysis of protein-protein interactions using a resonant mirror detection technique showed that 4.1R80 bound to Rch1 in vitro with high affinity (KD = 30 nM). The affinity decreased at least 7- and 20-fold, respectively, if the EED motif in exon 5 or if 4.1R NLS in exon 16 was lacking or mutated, confirming that both motifs were required for efficient importin-mediated nuclear import of 4.1R80.
Figures





References
-
- Amankwah KS, De Boni U. Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp Cell Res. 1994;210:315–325. - PubMed
-
- Anderson RA, Correas I, Mazzucco C, Castle JD, Marchesi VT. Tissue-specific analogues of erythrocyte protein 4.1 retain functional domains. J Cell Biochem. 1988;37:269–284. - PubMed
-
- Anderson RA, Lovrien RE. Glycophorin is linked by band 4.1 protein to the human erythrocyte membrane skeleton. Nature. 1984;307:655–658. - PubMed
-
- Ankenbauer T, Kleinschmidt JA, Walsh MJ, Weiner OH, Franke WW. Identification of a widespread nuclear actin binding protein. Nature. 1989;342:822–825. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

