Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion
- PMID: 10359599
- PMCID: PMC25377
- DOI: 10.1091/mbc.10.6.1821
Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion
Abstract
The amino acid sequence requirements of the transmembrane (TM) domain and cytoplasmic tail (CT) of the hemagglutinin (HA) of influenza virus in membrane fusion have been investigated. Fusion properties of wild-type HA were compared with those of chimeras consisting of the ectodomain of HA and the TM domain and/or CT of polyimmunoglobulin receptor, a nonviral integral membrane protein. The presence of a CT was not required for fusion. But when a TM domain and CT were present, fusion activity was greater when they were derived from the same protein than derived from different proteins. In fact, the chimera with a TM domain of HA and truncated CT of polyimmunoglobulin receptor did not support full fusion, indicating that the two regions are not functionally independent. Despite the fact that there is wide latitude in the sequence of the TM domain that supports fusion, a point mutation of a semiconserved residue within the TM domain of HA inhibited fusion. The ability of a foreign TM domain to support fusion contradicts the hypothesis that a pore is composed solely of fusion proteins and supports the theory that the TM domain creates fusion pores after a stage of hemifusion has been achieved.
Figures

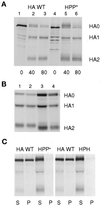
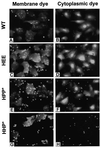
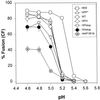
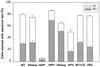
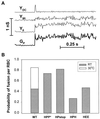
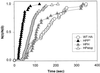
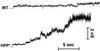
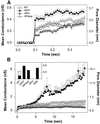


References
-
- Brown DA, London E. Structure of detergent-resistant membrane domains: does phase separation occur in biological membranes? Biochem Biophys Res Commun. 1997;240:1–7. - PubMed
-
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

