Activation of utrophin promoter by heregulin via the ets-related transcription factor complex GA-binding protein alpha/beta
- PMID: 10359616
- PMCID: PMC25417
- DOI: 10.1091/mbc.10.6.2075
Activation of utrophin promoter by heregulin via the ets-related transcription factor complex GA-binding protein alpha/beta
Abstract
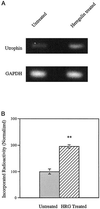
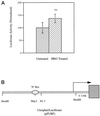
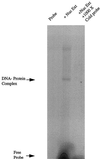
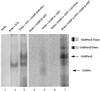
Utrophin/dystrophin-related protein is the autosomal homologue of the chromosome X-encoded dystrophin protein. In adult skeletal muscle, utrophin is highly enriched at the neuromuscular junction. However, the molecular mechanisms underlying regulation of utrophin gene expression are yet to be defined. Here we demonstrate that the growth factor heregulin increases de novo utrophin transcription in muscle cell cultures. Using mutant reporter constructs of the utrophin promoter, we define the N-box region of the promoter as critical for heregulin-mediated activation. Using this region of the utrophin promoter for DNA affinity purification, immunoblots, in vitro kinase assays, electrophoretic mobility shift assays, and in vitro expression in cultured muscle cells, we demonstrate that ets-related GA-binding protein alpha/beta transcription factors are activators of the utrophin promoter. Taken together, these results suggest that the GA-binding protein alpha/beta complex of transcription factors binds and activates the utrophin promoter in response to heregulin-activated extracellular signal-regulated kinase in muscle cell cultures. These findings suggest methods for achieving utrophin up-regulation in Duchenne's muscular dystrophy as well as mechanisms by which neurite-derived growth factors such as heregulin may influence the regulation of utrophin gene expression and subsequent enrichment at the neuromuscular junction of skeletal muscle.
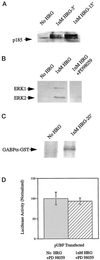
Figures




Similar articles
-
Induction of utrophin gene expression by heregulin in skeletal muscle cells: role of the N-box motif and GA binding protein.Proc Natl Acad Sci U S A. 1999 Mar 16;96(6):3223-7. doi: 10.1073/pnas.96.6.3223. Proc Natl Acad Sci U S A. 1999. PMID: 10077665 Free PMC article.
-
Sp1 and the ets-related transcription factor complex GABP alpha/beta functionally cooperate to activate the utrophin promoter.J Neurol Sci. 2002 May 15;197(1-2):27-35. doi: 10.1016/s0022-510x(02)00038-2. J Neurol Sci. 2002. PMID: 11997063
-
Ets-2 repressor factor silences extrasynaptic utrophin by N-box mediated repression in skeletal muscle.Mol Biol Cell. 2007 Aug;18(8):2864-72. doi: 10.1091/mbc.e06-12-1069. Epub 2007 May 16. Mol Biol Cell. 2007. PMID: 17507653 Free PMC article.
-
Multiple regulatory events controlling the expression and localization of utrophin in skeletal muscle fibers: insights into a therapeutic strategy for Duchenne muscular dystrophy.J Physiol Paris. 2002 Jan-Mar;96(1-2):31-42. doi: 10.1016/s0928-4257(01)00078-x. J Physiol Paris. 2002. PMID: 11755781 Review.
-
Regulation and functional significance of utrophin expression at the mammalian neuromuscular synapse.Microsc Res Tech. 2000 Apr 1;49(1):90-100. doi: 10.1002/(SICI)1097-0029(20000401)49:1<90::AID-JEMT10>3.0.CO;2-L. Microsc Res Tech. 2000. PMID: 10757882 Review.
Cited by
-
The ETS transcription factor GABPalpha is essential for early embryogenesis.Mol Cell Biol. 2004 Jul;24(13):5844-9. doi: 10.1128/MCB.24.13.5844-5849.2004. Mol Cell Biol. 2004. PMID: 15199140 Free PMC article.
-
Dystrophins, utrophins, and associated scaffolding complexes: role in mammalian brain and implications for therapeutic strategies.J Biomed Biotechnol. 2010;2010:849426. doi: 10.1155/2010/849426. Epub 2010 Jun 17. J Biomed Biotechnol. 2010. PMID: 20625423 Free PMC article. Review.
-
Single-nuclei sequencing of skeletal muscle reveals subsynaptic-specific transcripts involved in neuromuscular junction maintenance.Nat Commun. 2025 Mar 5;16(1):2220. doi: 10.1038/s41467-025-57487-1. Nat Commun. 2025. PMID: 40044687 Free PMC article.
-
Cis-Acting Sequence Elements and Upstream Open Reading Frame in Mouse Utrophin-A 5'-UTR Repress Cap-Dependent Translation.PLoS One. 2015 Jul 31;10(7):e0134809. doi: 10.1371/journal.pone.0134809. eCollection 2015. PLoS One. 2015. PMID: 26230628 Free PMC article.
-
Pathological pattern of Mdx mice diaphragm correlates with gradual expression of the short utrophin isoform Up71.Biochim Biophys Acta. 2006 Mar;1762(3):362-72. doi: 10.1016/j.bbadis.2005.11.006. Biochim Biophys Acta. 2006. PMID: 16457992 Free PMC article.
References
-
- Ausubel F, et al. Current Protocols in Molecular Biology. New York: Wiley; 1995.
-
- Brown RH. Dystrophin-associated proteins and muscular dystrophies. Annu Rev Med. 1997;48:457–466. - PubMed
-
- Brown TA, McKnight SL. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes & Dev. 1992;6:2502–2512. - PubMed
-
- Campbell KP. Three muscular dystrophies: loss of cytoskeletal extracellular matrix linkage. Cell. 1996;80:675–679. - PubMed
-
- Campbell KP, Crosbie RH. Muscular dystrophy. Utrophin to the rescue. Nature. 1996;384:308–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

