Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease
- PMID: 10364314
- PMCID: PMC112623
- DOI: 10.1128/JVI.73.7.5646-5653.1999
Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease
Abstract
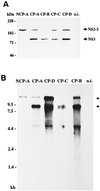
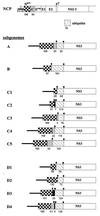
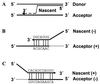
Four bovine viral diarrhea virus type 2 (BVDV-2) pairs consisting of cytopathogenic (cp) and noncp BVDV-2 were isolated during an outbreak of mucosal disease. Comparative sequence analysis showed that the four noncp BVDV-2 isolates were almost identical. For the cp BVDV-2 isolates, viral subgenomic RNAs were shown by Northern blot to have a length of about 8 kb, which is about 4.3 kb shorter than the genome of noncp BVDV. Cytopathogenicity and the expression of NS3 were both strictly correlated to the presence of viral subgenomic RNAs. By reverse transcription-PCR, Southern blot analysis, and nucleotide sequencing, a set of 11 unique subgenomes was identified with up to 5 different subgenomes isolated from one animal. To our knowledge, this is the first report on isolation of a set of pestiviral subgenomes from individual animals. Common features of the BVDV-2 subgenomic RNAs include (i) deletion of most of the genomic region encoding the structural proteins, as well as the nonstructural proteins p7 and NS2, and (ii) insertion of cellular (poly)ubiquitin coding sequences. Three subgenomes also comprised 15 to 75 nucleotides derived from the 5' part of the NS2 gene. Comparisons of the obtained nucleotide sequences revealed that the different BVDV-2 subgenomes evolved from the respective noncp BVDV-2 by RNA recombination. The presence of short regions of sequence similarity at several crossing-over sites suggests that base pairing between the nascent RNA strand and the acceptor RNA template facilitates template switching of the BVDV RNA-dependent RNA polymerase.
Figures


References
-
- Agol V I. Recombination and other genomic rearrangements in picornaviruses. Semin Virol. 1997;8:77–84.
-
- Becher P, König M, Paton D, Thiel H-J. Further characterization of border disease virus isolates: evidence for the presence of more than three species within the genus pestivirus. Virology. 1995;209:200–206. - PubMed
-
- Becher P, Orlich M, Shannon A D, Horner G, König M, Thiel H-J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol. 1997;78:1357–1366. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

