Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription
- PMID: 10364319
- PMCID: PMC112628
- DOI: 10.1128/JVI.73.7.5688-5697.1999
Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription
Abstract
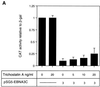
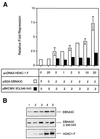
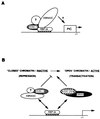
EBNA3C can specifically repress the expression of reporter plasmids containing EBV Cp latency-associated promoter elements. Cp is normally the main promoter for EBNA mRNA initiation, so it appears that EBNA3C contributes to a negative autoregulatory control loop. By mutational analysis it was previously established that this repression is consistent with EBNA3C being targeted to Cp by binding the cellular sequence-specific DNA-binding protein CBF1 (also known as recombination signal-binding protein [RBP]-Jkappa. Further analysis suggested that in vivo a corepressor interacts with EBNA3C in this DNA binding complex. Results presented here are all consistent with a component of such a corepressor exhibiting histone deacetylase activity. The drug trichostatin A, which specifically inhibits histone deacetylases, relieved two- to threefold the repression of Cp induced by EBNA3C in two different cell types. Moreover, repression of pTK-CAT-Cp4x by EBNA3C was specifically enhanced by cotransfection of an expression plasmid for human histone deacetylase-1 (HDAC1). Consistent with these functional assays, in vitro-translated HDAC1 bound to a glutathione S-transferase (GST) fusion protein including full-length EBNA3C, and in the reciprocal experiment EBNA3C bound to a GST fusion with the N terminus of HDAC1. Coimmunoprecipitations also revealed an EBNA3C-HDAC1 interaction in vivo, and GST-EBNA3C bound functional histone deacetylase enzyme activity from HeLa cell nuclear extracts. The region of EBNA3C involved in the interaction with HDAC1 appears to correspond to the region which is necessary for binding to CBF1/RBP-Jkappa. A direct physical interaction between EBNA3C and HDAC1 was demonstrated with recombinant proteins purified from bacterial cells, and we therefore conclude that HDAC1 and CBF1/RBP-Jkappa bind to the same or adjacent regions of EBNA3C. These data suggest that recruitment of histone deacetylase activity makes a significant contribution to the repression of transcription from Cp because EBNA3C bridges an interaction between CBF1/RBP-Jkappa and HDAC1.
Figures








Similar articles
-
Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21.J Virol. 1997 Nov;71(11):8552-62. doi: 10.1128/JVI.71.11.8552-8562.1997. J Virol. 1997. PMID: 9343213 Free PMC article.
-
Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines.J Virol. 2003 Apr;77(7):4261-72. doi: 10.1128/jvi.77.7.4261-4272.2003. J Virol. 2003. PMID: 12634383 Free PMC article.
-
Transcriptional repression by the Epstein-Barr virus EBNA3A protein tethered to DNA does not require RBP-Jkappa.J Gen Virol. 1998 Feb;79 ( Pt 2):363-70. doi: 10.1099/0022-1317-79-2-363. J Gen Virol. 1998. PMID: 9472621
-
The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration.Front Biosci. 2002 Mar 1;7:d704-16. doi: 10.2741/subraman. Front Biosci. 2002. PMID: 11861219 Review.
-
Chemical genetics: exploring and controlling cellular processes with chemical probes.Trends Biochem Sci. 1999 Aug;24(8):317-20. doi: 10.1016/s0968-0004(99)01425-5. Trends Biochem Sci. 1999. PMID: 10431176 Review.
Cited by
-
The expression and function of Epstein-Barr virus encoded latent genes.Mol Pathol. 2000 Oct;53(5):238-47. doi: 10.1136/mp.53.5.238. Mol Pathol. 2000. PMID: 11091847 Free PMC article. Review.
-
A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome.EMBO J. 2001 May 15;20(10):2367-75. doi: 10.1093/emboj/20.10.2367. EMBO J. 2001. PMID: 11350925 Free PMC article.
-
Epstein-Barr Virus History and Pathogenesis.Viruses. 2023 Mar 9;15(3):714. doi: 10.3390/v15030714. Viruses. 2023. PMID: 36992423 Free PMC article. Review.
-
Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation.Nucleic Acids Res. 2000 Oct 15;28(20):3918-25. doi: 10.1093/nar/28.20.3918. Nucleic Acids Res. 2000. PMID: 11024171 Free PMC article.
-
BCL11B is a general transcriptional repressor of the HIV-1 long terminal repeat in T lymphocytes through recruitment of the NuRD complex.Virology. 2008 Oct 25;380(2):173-81. doi: 10.1016/j.virol.2008.07.035. Epub 2008 Sep 2. Virology. 2008. PMID: 18768194 Free PMC article.
References
-
- Alfieri C, Birckenbach M, Kieff E. Early events in Epstein-Barr virus infection of human lymphocytes. Virology. 1991;181:595–608. - PubMed
-
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- Allday M J, Crawford D H, Griffin B E. Epstein-Barr virus latent gene expression during the initiation of B cell immortalisation. J Gen Virol. 1989;70:1755–1764. - PubMed
-
- Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

