Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription
- PMID: 10364337
- PMCID: PMC112646
- DOI: 10.1128/JVI.73.7.5852-5864.1999
Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription
Abstract
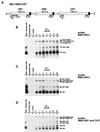
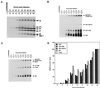
M2-1 protein of human respiratory syncytial virus (RSV) is a transcription antitermination factor that is important for the efficient synthesis of full-length mRNAs as well as for the synthesis of polycistronic readthrough mRNAs, which are characteristic of nonsegmented negative-strand RNA viruses. The contributions of these effects to RSV sequential transcription were investigated with minigenomes which contained one to five genes which were either foreign marker genes or authentic RSV genes. When evaluated on a promoter-proximal gene, the effect of M2-1 on the synthesis of full-length mRNA was much greater for a long (1,212- or 1,780-nucleotide) gene (up to a 615-fold increase) than for a short (274-nucleotide) gene (less than a 2-fold increase). This was independent of whether the gene contained non-RSV or RSV-specific sequence. Once the polymerase had terminated prematurely, it was unable to reinitiate at a downstream gene. These studies also confirmed that M2-1 enhances the synthesis of polycistronic mRNAs and that the magnitude of this effect varied greatly among different naturally occurring gene junctions. The synthesis of polycistronic mRNAs, which presumably involves antitermination at the gene-end signal, required a higher level of M2-1 than did the synthesis of the corresponding monocistronic mRNAs. M2-1 did not have a comparable antitermination effect at the junction between the leader region and the first gene. In a minigenome containing the NS1 and NS2 genes in their authentic sequence context, synthesis of full-length NS1 and NS2 mRNAs in the absence of M2-1 was remarkably high (36 and 57%, respectively, of the maximum levels observed in the presence of M2-1). In contrast, synthesis of mRNA from additional downstream genes was highly dependent on M2-1. Thus, RSV has the potential for two transcription programs: one in the absence of M2-1, in which only the NS1 and NS2 genes are transcribed, and one in the presence of M2-1, in which sequential transcription of the complete genome occurs. The dependence on M2-1 for transcription was greater for a gene in the fifth position from the promoter than for one in the third position. This indicates that under conditions where M2-1 is limiting, its concentration affects the gradient of transcription. Although M2-1 was found to have profound effects on transcription, it had no effect on replication of any minigenome tested, suggesting that it is not an active participant in RNA replication or regulation of RNA replication. Finally, since a permissive RSV infection is marked by a gradual increase in the intracellular accumulation of viral proteins including M2-1, we examined the relative abundances of various mRNAs during RSV infection for evidence of temporal regulation of transcription. None was found, implying that the availability of M2-1 during a permissive infection is sufficient at all times such that its concentration does not mediate temporal regulation of gene transcription.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

