Hepatitis A virus capsid protein VP1 has a heterogeneous C terminus
- PMID: 10364353
- PMCID: PMC112662
- DOI: 10.1128/JVI.73.7.6015-6023.1999
Hepatitis A virus capsid protein VP1 has a heterogeneous C terminus
Abstract
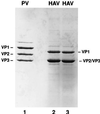
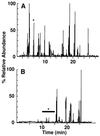
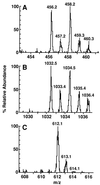
Hepatitis A virus (HAV) encodes a single polyprotein which is posttranslationally processed into the functional structural and nonstructural proteins. Only one protease, viral protease 3C, has been implicated in the nine protein scissions. Processing of the capsid protein precursor region generates a unique intermediate, PX (VP1-2A), which accumulates in infected cells and is assumed to serve as precursor to VP1 found in virions, although the details of this reaction have not been determined. Coexpression in transfected cells of a variety of P1 precursor proteins with viral protease 3C demonstrated efficient production of PX, as well as VP0 and VP3; however, no mature VP1 protein was detected. To identify the C-terminal amino acid residue of HAV VP1, we performed peptide sequence analysis by protease-catalyzed [18O]H2O incorporation followed by liquid chromatography ion-trap microspray tandem mass spectrometry of HAV VP1 isolated from purified virions. Two different cell culture-adapted isolates of HAV, strains HM175pE and HM175p35, were used for these analyses. VP1 preparations from both virus isolates contained heterogeneous C termini. The predominant C-terminal amino acid in both virus preparations was VP1-Ser274, which is located N terminal to a methionine residue in VP1-2A. In addition, the analysis of HM175pE recovered smaller amounts of amino acids VP1-Glu273 and VP1-Thr272. In the case of HM175p35, which contains valine at amino acid position VP1-273, VP1-Thr272 was found in addition to VP1-Ser274. The data suggest that HAV 3C is not the protease responsible for generation of the VP1 C terminus. We propose the involvement of host cell protease(s) in the production of HAV VP1.
Figures


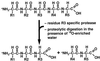



References
-
- Bishop N E, Anderson D A. Hepatitis A virus subviral particles: purification, accumulation, and relative infectivity of virions, provirions and procapsids. Arch Virol. 1997;142:2147–2160. - PubMed
-
- Boege U, Scraba D G. Mengo virus maturation is accompanied by C-terminal modification of capsid protein VP1. Virology. 1989;168:409–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

