Identification of a cis-regulatory element involved in phytochrome down-regulated expression of the pea small GTPase gene pra2
- PMID: 10364400
- PMCID: PMC59287
- DOI: 10.1104/pp.120.2.491
Identification of a cis-regulatory element involved in phytochrome down-regulated expression of the pea small GTPase gene pra2
Abstract
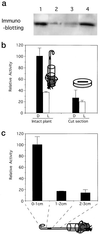
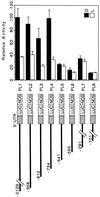
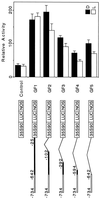
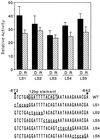
The pra2 gene encodes a pea (Pisum sativum) small GTPase belonging to the YPT/rab family, and its expression is down-regulated by light, mediated by phytochrome. We have isolated and characterized a genomic clone of this gene and constructed a fusion DNA of its 5'-upstream region in front of the gene for firefly luciferase. Using this construct in a transient assay, we determined a pra2 cis-regulatory region sufficient to direct the light down-regulation of the luciferase reporter gene. Both 5'- and internal deletion analyses revealed that the 93-bp sequence between -734 and -642 from the transcriptional start site was important for phytochrome down-regulation. Gain-of-function analysis showed that this 93-bp region could confer light down-regulation when fused to the cauliflower mosaic virus 35S promoter. Furthermore, linker-scanning analysis showed that a 12-bp sequence within the 93-bp region mediated phytochrome down-regulation. Gel-retardation analysis showed the presence of a nuclear factor that was specifically bound to the 12-bp sequence in vitro. These results indicate that this element is a cis-regulatory element involved in phytochrome down-regulated expression.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

