Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli
- PMID: 10368138
- PMCID: PMC93841
- DOI: 10.1128/JB.181.12.3658-3665.1999
Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli
Abstract
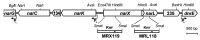
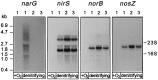
Bacterial denitrification is expressed in response to the concurrent exogenous signals of low-oxygen tension and nitrate or one of its reduction products. The mechanism by which nitrate-dependent gene activation is effected was investigated in the denitrifying bacterium Pseudomonas stutzeri ATCC 14405. We have identified and isolated from this organism the chromosomal region encoding the two-component sensor-regulator pair NarXL and found that it is linked with the narG operon for respiratory nitrate reductase. The same region encodes two putative nitrate or nitrite translocases, NarK and NarC (the latter shows the highest similarity to yeast [Pichia] and plant [Nicotiana] nitrate transporters), and the nitrate-regulated transcription factor, DnrE, of the FNR family. The roles of NarX and NarL in nitrate respiration were studied with deletion mutants. NarL activated the transcription of narG, narK, and dnrE but did not affect the denitrification regulons for the respiratory substrates nitrite, nitric oxide, and nitrous oxide. The promoters of narG, narK, and dnrE carry sequence motifs, TACYYMT, which correspond to the NarL recognition sequence established for Escherichia coli. The cellular response toward nitrate and nitrite was mediated by the sensor protein NarX, which discriminated weakly between these oxyanions. Our data show that the NarXL two-component regulatory system has been incorporated into the bacterial denitrification process of P. stutzeri for selective regulation of nitrate respiration.
Figures




Similar articles
-
'Locked-on' and 'locked-off' signal transduction mutations in the periplasmic domain of the Escherichia coli NarQ and NarX sensors affect nitrate- and nitrite-dependent regulation by NarL and NarP.Mol Microbiol. 1997 Jun;24(5):1049-60. doi: 10.1046/j.1365-2958.1997.4131779.x. Mol Microbiol. 1997. PMID: 9220011
-
Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeri during the transition from aerobic respiration to denitrification: evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system.J Bacteriol. 1999 Jan;181(1):161-6. doi: 10.1128/JB.181.1.161-166.1999. J Bacteriol. 1999. PMID: 9864326 Free PMC article.
-
Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons.Microbiology (Reading). 1999 Jan;145 ( Pt 1):41-55. doi: 10.1099/13500872-145-1-41. Microbiology (Reading). 1999. PMID: 10206709
-
Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli.Mol Microbiol. 1993 Aug;9(3):425-34. doi: 10.1111/j.1365-2958.1993.tb01704.x. Mol Microbiol. 1993. PMID: 8412692 Review.
-
Nitrate and nitrite transport in bacteria.Cell Mol Life Sci. 2001 Feb;58(2):215-24. doi: 10.1007/PL00000849. Cell Mol Life Sci. 2001. PMID: 11289303 Free PMC article. Review.
Cited by
-
Aerobic Denitrification and Heterotrophic Sulfur Oxidation in the Genus Halomonas Revealed by Six Novel Species Characterizations and Genome-Based Analysis.Front Microbiol. 2021 Mar 18;12:652766. doi: 10.3389/fmicb.2021.652766. eCollection 2021. Front Microbiol. 2021. PMID: 33815342 Free PMC article.
-
Control of gene expression by FNR-like proteins in facultatively anaerobic bacteria.Folia Microbiol (Praha). 2002;47(2):95-103. doi: 10.1007/BF02817665. Folia Microbiol (Praha). 2002. PMID: 12058404 Review.
-
Absence of 4-Formylaminooxyvinylglycine Production by Pseudomonas fluorescens WH6 Results in Resource Reallocation from Secondary Metabolite Production to Rhizocompetence.Microorganisms. 2021 Mar 31;9(4):717. doi: 10.3390/microorganisms9040717. Microorganisms. 2021. PMID: 33807194 Free PMC article.
-
The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration.J Bacteriol. 2007 Jun;189(11):4310-4. doi: 10.1128/JB.00240-07. Epub 2007 Mar 30. J Bacteriol. 2007. PMID: 17400734 Free PMC article.
-
Maximal expression of membrane-bound nitrate reductase in Paracoccus is induced by nitrate via a third FNR-like regulator named NarR.J Bacteriol. 2001 Jun;183(12):3606-13. doi: 10.1128/JB.183.12.3606-3613.2001. J Bacteriol. 2001. PMID: 11371524 Free PMC article.
References
-
- Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli. J Biol Chem. 1981;256:11905–11910. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995.
-
- Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Cascio D, Gunsalus R P, Dickerson R E. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry. 1998;37:3665–3676. - PubMed
-
- Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coliresponse regulator NarL. Biochemistry. 1996;35:11053–11061. - PubMed
-
- Blasco F, Iobbi C, Giordano G, Chippaux M, Bonnefoy V. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the naroperon and reassessment of the role of the α and β subunits in iron binding and electron transfer. Mol Gen Genet. 1989;218:249–256. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

