The ATP-dependent HslVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli
- PMID: 10368140
- PMCID: PMC93843
- DOI: 10.1128/JB.181.12.3674-3680.1999
The ATP-dependent HslVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli
Abstract
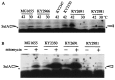
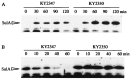
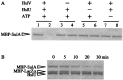
Escherichia coli mutants lacking activities of all known cytosolic ATP-dependent proteases (Lon, ClpAP, ClpXP, and HslVU), due to double deletions [DeltahslVU and Delta(clpPX-lon)], cannot grow at low (30 degrees C) or very high (45 degrees C) temperatures, unlike those carrying either of the deletions. Such growth defects were particularly marked when the deletions were introduced into strain MG1655 or W3110. To examine the functions of HslVU and other proteases further, revertants that can grow at 30 degrees C were isolated from the multiple-protease mutant and characterized. The revertants were found to carry a suppressor affecting either ftsZ (encoding a key cell division protein) or sulA (encoding the SulA inhibitor, which binds and inhibits FtsZ). Whereas the ftsZ mutations were identical to a mutation known to produce a protein refractory to SulA inhibition, the sulA mutations affected the promoter-operator region, reducing synthesis of SulA. These results suggested that the growth defect of the parental double-deletion mutant at a low temperature was due to the accumulation of excess SulA without DNA-damaging treatment. Consistent with these results, SulA in the double-deletion mutant was much more stable than that in the Delta(clpPX-lon) mutant, suggesting that SulA can be degraded by HslVU. As expected, purified HslVU protease degraded SulA (fused to the maltose-binding protein) efficiently in an ATP-dependent manner. These results suggest that HslVU as well as Lon participates in the in vivo turnover of SulA and that HslVU becomes essential for growth when the Lon (and Clp) protease level is reduced below a critical threshold.
Figures


Similar articles
-
ATP-dependent degradation of SulA, a cell division inhibitor, by the HslVU protease in Escherichia coli.FEBS Lett. 1999 Jul 30;456(1):211-4. doi: 10.1016/s0014-5793(99)00935-7. FEBS Lett. 1999. PMID: 10452560
-
Functional dissection of a cell-division inhibitor, SulA, of Escherichia coli and its negative regulation by Lon.Mol Gen Genet. 1997 Apr 28;254(4):351-7. doi: 10.1007/s004380050426. Mol Gen Genet. 1997. PMID: 9180687
-
Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of sigma32 and abnormal proteins in Escherichia coli.J Bacteriol. 1997 Dec;179(23):7219-25. doi: 10.1128/jb.179.23.7219-7225.1997. J Bacteriol. 1997. PMID: 9393683 Free PMC article.
-
[Structural and functional characteristics of ATP-dependent Lon-proteinase from Escherichia coli].Bioorg Khim. 1999 Dec;25(12):883-91. Bioorg Khim. 1999. PMID: 10734549 Review. Russian.
-
ATP-dependent proteases that also chaperone protein biogenesis.Trends Biochem Sci. 1997 Apr;22(4):118-23. doi: 10.1016/s0968-0004(97)01020-7. Trends Biochem Sci. 1997. PMID: 9149530 Review.
Cited by
-
Analysis of the Escherichia coli Alp phenotype: heat shock induction in ssrA mutants.J Bacteriol. 2005 Jul;187(14):4739-51. doi: 10.1128/JB.187.14.4739-4751.2005. J Bacteriol. 2005. PMID: 15995188 Free PMC article.
-
Checkpoints That Regulate Balanced Biosynthesis of Lipopolysaccharide and Its Essentiality in Escherichia coli.Int J Mol Sci. 2021 Dec 24;23(1):189. doi: 10.3390/ijms23010189. Int J Mol Sci. 2021. PMID: 35008618 Free PMC article. Review.
-
A Transient π-π or Cation-π Interaction between Degron and Degrader Dual Residues: A Key Step for the Substrate Recognition and Discrimination in the Processive Degradation of SulA by ClpYQ (HslUV) Protease in Escherichia coli.Int J Mol Sci. 2023 Dec 11;24(24):17353. doi: 10.3390/ijms242417353. Int J Mol Sci. 2023. PMID: 38139184 Free PMC article.
-
Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site.Proc Natl Acad Sci U S A. 2010 Mar 2;107(9):4028-33. doi: 10.1073/pnas.1000315107. Epub 2010 Feb 16. Proc Natl Acad Sci U S A. 2010. PMID: 20160114 Free PMC article.
-
Discovery of a dual protease mechanism that promotes DNA damage checkpoint recovery.PLoS Genet. 2018 Jul 6;14(7):e1007512. doi: 10.1371/journal.pgen.1007512. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 29979679 Free PMC article.
References
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Bukau B. Regulation of the Escherichia coliheat-shock response. Mol Microbiol. 1993;9:671–680. - PubMed
-
- Chuang S-E, Burland V, Plunkett III G, Daniels D L, Blattner F R. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene. 1993;134:1–6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

