Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa
- PMID: 10368148
- PMCID: PMC93851
- DOI: 10.1128/JB.181.12.3730-3742.1999
Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa
Abstract
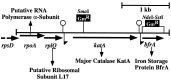
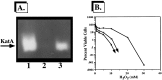
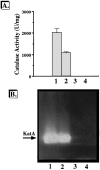
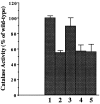
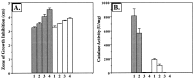
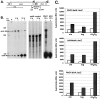
We have cloned a 3.6-kb genomic DNA fragment from Pseudomonas aeruginosa harboring the rpoA, rplQ, katA, and bfrA genes. These loci are predicted to encode, respectively, (i) the alpha subunit of RNA polymerase; (ii) the L17 ribosomal protein; (iii) the major catalase, KatA; and (iv) one of two iron storage proteins called bacterioferritin A (BfrA; cytochrome b1 or b557). Our goal was to determine the contributions of KatA and BfrA to the resistance of P. aeruginosa to hydrogen peroxide (H2O2). When provided on a multicopy plasmid, the P. aeruginosa katA gene complemented a catalase-deficient strain of Escherichia coli. The katA gene was found to contain two translational start codons encoding a heteromultimer of approximately 160 to 170 kDa and having an apparent Km for H2O2 of 44.7 mM. Isogenic katA and bfrA mutants were hypersusceptible to H2O2, while a katA bfrA double mutant demonstrated the greatest sensitivity. The katA and katA bfrA mutants possessed no detectable catalase activity. Interestingly, a bfrA mutant expressed only approximately 47% the KatA activity of wild-type organisms, despite possessing wild-type katA transcription and translation. Plasmids harboring bfrA genes encoding BfrA altered at critical amino acids essential for ferroxidase activity could not restore wild-type catalase activity in the bfrA mutant. RNase protection assays revealed that katA and bfrA are on different transcripts, the levels of which are increased by both iron and H2O2. Mass spectrometry analysis of whole cells revealed no significant difference in total cellular iron levels in the bfrA, katA, and katA bfrA mutants relative to wild-type bacteria. Our results suggest that P. aeruginosa BfrA may be required as one source of iron for the heme prosthetic group of KatA and thus for protection against H2O2.
Figures


Similar articles
-
Iron Mineralizing Bacterioferritin A from Mycobacterium tuberculosis Exhibits Unique Catalase-Dps-like Dual Activities.Inorg Chem. 2019 Apr 15;58(8):4741-4752. doi: 10.1021/acs.inorgchem.8b02758. Epub 2019 Mar 28. Inorg Chem. 2019. PMID: 30920210
-
Neisseria gonorrhoeae bacterioferritin: structural heterogeneity, involvement in iron storage and protection against oxidative stress.Microbiology (Reading). 1999 Oct;145 ( Pt 10):2967-75. doi: 10.1099/00221287-145-10-2967. Microbiology (Reading). 1999. PMID: 10537219
-
Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide.J Bacteriol. 1995 Nov;177(22):6536-44. doi: 10.1128/jb.177.22.6536-6544.1995. J Bacteriol. 1995. PMID: 7592431 Free PMC article.
-
Iron storage in bacteria.Adv Microb Physiol. 1998;40:281-351. doi: 10.1016/s0065-2911(08)60134-4. Adv Microb Physiol. 1998. PMID: 9889981 Review.
-
Bacterioferritin: a hemoprotein member of the ferritin family.Adv Exp Med Biol. 1994;356:157-64. doi: 10.1007/978-1-4615-2554-7_18. Adv Exp Med Biol. 1994. PMID: 7887220 Review. No abstract available.
Cited by
-
Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment.Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):18247-52. doi: 10.1073/pnas.0706739104. Epub 2007 Nov 7. Proc Natl Acad Sci U S A. 2007. PMID: 17989226 Free PMC article.
-
Iron storage proteins are essential for the survival and pathogenesis of Mycobacterium tuberculosis in THP-1 macrophages and the guinea pig model of infection.J Bacteriol. 2012 Feb;194(3):567-75. doi: 10.1128/JB.05553-11. Epub 2011 Nov 18. J Bacteriol. 2012. PMID: 22101841 Free PMC article.
-
Abiotic and biotic factors responsible for antimonite oxidation in Agrobacterium tumefaciens GW4.Sci Rep. 2017 Mar 2;7:43225. doi: 10.1038/srep43225. Sci Rep. 2017. PMID: 28252030 Free PMC article.
-
A Critical Review of Resistance and Oxidation Mechanisms of Sb-Oxidizing Bacteria for the Bioremediation of Sb(III) Pollution.Front Microbiol. 2021 Sep 7;12:738596. doi: 10.3389/fmicb.2021.738596. eCollection 2021. Front Microbiol. 2021. PMID: 34557178 Free PMC article. Review.
-
Functional Characterization of Alr0765, A Hypothetical Protein from Anabaena PCC 7120 Involved in Cellular Energy Status Sensing, Iron Acquisition and Abiotic Stress Management in E. coli Using Molecular, Biochemical and Computational Approaches.Curr Genomics. 2020 May;21(4):295-310. doi: 10.2174/1389202921999200424181239. Curr Genomics. 2020. PMID: 33071622 Free PMC article.
References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andrews S C, Smith J M, Yewdall S J, Guest J R, Harrison P M. Bacterioferritins and ferritins are distantly related in evolution: conservation of ferroxidase-centre residues. FEBS Lett. 1991;293:164–168. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. pp. 5.3.2–5.3.8.
-
- Bando Y, Aki K. Superoxide-mediated release of iron from ferritin by some flavoenzymes. Biochem Biophys Res Commun. 1990;168:389–395. - PubMed
-
- Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

