Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks
- PMID: 10368149
- PMCID: PMC93852
- DOI: 10.1128/JB.181.12.3743-3750.1999
Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks
Abstract
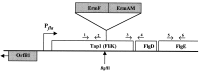
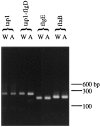
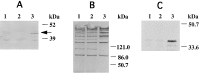
The treponemal fla operon is comprised of numerous motility-related genes; however, the initial gene of this operon, tap1, has no known function. A recently developed system to generate specific mutants in Treponema denticola was utilized to determine if Tap1 was essential for motility. T. denticola tap1 and flanking DNA were identified, cloned, and sequenced, and a suicide plasmid that contained tap1 interrupted with an erythromycin resistance cassette (ermF and ermAM) was constructed. Because of potential polar effects from this cassette, a second plasmid that contained tap1 interrupted with a modified erythromycin resistance cassette that lacked the putative ermF transcription terminator was constructed. Electroporation-mediated allelic exchange incorporated the interrupted tap1 genes into the T. denticola chromosome, creating Tap1-deficient mutants. Reverse transcriptase PCR revealed that the erythromycin resistance cassette within tap1 did not terminate fla operon transcription in either mutant. Moreover, the phenotypes of the two mutants were indistinguishable. These mutants lacked motion in liquid culture, were unable to spread on agar plates, and lacked flagellar filaments as determined by electron microscopy. Immunoblots revealed a marked reduction in detectable FlaB flagellar filament protein compared to that of wild type; however, flaB RNA was easily detectable, and transcription levels did not appear to be altered. The basis for the lack of filament protein expression is unknown. Immunoblotting also showed that the flagellar hook protein (FlgE) was synthesized in the Tap1-deficient mutant; however, electron microscopy revealed that the mutant possessed unusual elongated hooks of variable lengths. We propose that treponemal Tap1 is analogous to FliK, which is involved in monitoring the flagellar hook length of Salmonella typhimurium.
Figures






Similar articles
-
Molecular characterization of a flagellar (fla) operon in the oral spirochete Treponema denticola ATCC 35405.FEMS Microbiol Lett. 1999 Oct 1;179(1):31-6. doi: 10.1111/j.1574-6968.1999.tb08703.x. FEMS Microbiol Lett. 1999. PMID: 10481082
-
Genetic and biochemical analysis of the flagellar hook of Treponema phagedenis.J Bacteriol. 1994 Jun;176(12):3631-7. doi: 10.1128/jb.176.12.3631-3637.1994. J Bacteriol. 1994. PMID: 8206841 Free PMC article.
-
Organization, transcription, and expression of the 5' region of the fla operon of Treponema phagedenis and Treponema pallidum.J Bacteriol. 1996 Aug;178(15):4628-34. doi: 10.1128/jb.178.15.4628-4634.1996. J Bacteriol. 1996. PMID: 8755894 Free PMC article.
-
Genetic and transcriptional analysis of flgB flagellar operon constituents in the oral spirochete Treponema denticola and their heterologous expression in enteric bacteria.Infect Immun. 1997 Jun;65(6):2041-51. doi: 10.1128/iai.65.6.2041-2051.1997. Infect Immun. 1997. PMID: 9169730 Free PMC article.
-
The FliK protein and flagellar hook-length control.Protein Sci. 2007 May;16(5):769-80. doi: 10.1110/ps.072785407. Protein Sci. 2007. PMID: 17456739 Free PMC article. Review.
Cited by
-
Inactivation of cyclic Di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola.J Bacteriol. 2013 Sep;195(17):3897-905. doi: 10.1128/JB.00610-13. Epub 2013 Jun 21. J Bacteriol. 2013. PMID: 23794624 Free PMC article.
-
Cloning and expression of two novel hemin binding protein genes from Treponema denticola.Infect Immun. 2001 Jul;69(7):4465-72. doi: 10.1128/IAI.69.7.4465-4472.2001. Infect Immun. 2001. PMID: 11401987 Free PMC article.
-
A pleiotropic role of FlaG in regulating the cell morphogenesis and flagellar homeostasis at the cell poles of Treponema denticola.Cell Microbiol. 2019 Feb;21(2):e12886. doi: 10.1111/cmi.12886. Epub 2018 Jul 23. Cell Microbiol. 2019. PMID: 29935042 Free PMC article.
-
Initial characterization of the FlgE hook high molecular weight complex of Borrelia burgdorferi.PLoS One. 2014 May 23;9(5):e98338. doi: 10.1371/journal.pone.0098338. eCollection 2014. PLoS One. 2014. PMID: 24859001 Free PMC article.
-
Evidence that TP_0144 of Treponema pallidum is a thiamine-binding protein.J Bacteriol. 2015 Apr;197(7):1164-72. doi: 10.1128/JB.02472-14. Epub 2015 Jan 20. J Bacteriol. 2015. PMID: 25605310 Free PMC article.
References
-
- Compton T. Degenerate primers for DNA amplification. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 39–45.
-
- Dantuono L A. Characterization of the fla motility operon of Treponema phagedenis. M.S. thesis. Albany, N.Y: State University of New York; 1996.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

