Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons
- PMID: 10368156
- PMCID: PMC93859
- DOI: 10.1128/JB.181.12.3803-3809.1999
Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons
Abstract
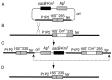
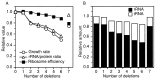
The Escherichia coli genome carries seven rRNA (rrn) operons, each containing three rRNA genes. The presence of multiple operons has been an obstacle to many studies of rRNA because the effect of mutations in one operon is diluted by the six remaining wild-type copies. To create a tool useful for manipulating rRNA, we sequentially inactivated from one to all seven of these operons with deletions spanning the 16S and 23S rRNA genes. In the final strain, carrying no intact rRNA operon on the chromosome, rRNA molecules were expressed from a multicopy plasmid containing a single rRNA operon (prrn). Characterization of these rrn deletion strains revealed that deletion of two operons was required to observe a reduction in the growth rate and rRNA/protein ratio. When the number of deletions was extended from three to six, the decrease in the growth rate was slightly more than the decrease in the rRNA/protein ratio, suggesting that ribosome efficiency was reduced. This reduction was most pronounced in the Delta7 prrn strain, in which the growth rate, unlike the rRNA/protein ratio, was not completely restored to wild-type levels by a cloned rRNA operon. The decreases in growth rate and rRNA/protein ratio were surprisingly moderate in the rrn deletion strains; the presence of even a single operon on the chromosome was able to produce as much as 56% of wild-type levels of rRNA. We discuss possible applications of these strains in rRNA studies.
Figures
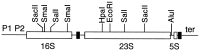


References
-
- Beresford T, Condon S. Cloning and partial characterization of genes for ribonucleic acid in Lactococcus lactis subsp. lactis. FEMS Microbiol Lett. 1991;62:319–323. - PubMed
-
- Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacBgene and a temperature-sensitive pSC101 replicon. Mol Microbiol. 1991;5:1447–1457. - PubMed
-
- Bott K, Stewart G C, Anderson A G. Genetic mapping of cloned ribosomal RNA genes. In: Hoch J A, Ganesan A T, editors. Syntro Conference on Genetics and Biotechnology of Bacilli. New York, N.Y: Academic Press; 1984. pp. 19–34.
-
- Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 1553–1569.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

