Activation of Escherichia coli leuV transcription by FIS
- PMID: 10368169
- PMCID: PMC93872
- DOI: 10.1128/JB.181.12.3864-3868.1999
Activation of Escherichia coli leuV transcription by FIS
Abstract
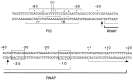
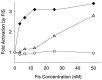
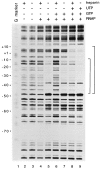
The transcription factor FIS has been implicated in the regulation of several stable RNA promoters, including that for the major tRNALeu species in Escherichia coli, tRNA1Leu. However, no evidence for direct involvement of FIS in tRNA1Leu expression has been reported. We show here that FIS binds to a site upstream of the leuV promoter (centered at -71) and that it directly stimulates leuV transcription in vitro. A mutation in the FIS binding site reduces transcription from a leuV promoter in strains containing FIS but has no effect on transcription in strains lacking FIS, indicating that FIS contributes to leuV expression in vivo. We also find that RNA polymerase forms an unusual heparin-sensitive complex with the leuV promoter, having a downstream protection boundary of approximately -7, and that the first two nucleotides of the transcript, GTP and UTP, are required for formation of a heparin-stable complex that extends downstream of the transcription start site. These studies have implications for the regulation of leuV transcription.
Figures

Similar articles
-
Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism.Mol Microbiol. 2004 Jul;53(2):665-74. doi: 10.1111/j.1365-2958.2004.04147.x. Mol Microbiol. 2004. PMID: 15228542
-
Multiple mechanisms are used for growth rate and stringent control of leuV transcriptional initiation in Escherichia coli.J Bacteriol. 1999 Sep;181(18):5771-82. doi: 10.1128/JB.181.18.5771-5782.1999. J Bacteriol. 1999. PMID: 10482520 Free PMC article.
-
Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis.J Bacteriol. 1997 Oct;179(20):6367-77. doi: 10.1128/jb.179.20.6367-6377.1997. J Bacteriol. 1997. PMID: 9335285 Free PMC article.
-
DNA supercoiling and transcription in Escherichia coli: The FIS connection.Biochimie. 2001 Feb;83(2):213-7. doi: 10.1016/s0300-9084(00)01217-7. Biochimie. 2001. PMID: 11278071 Review.
-
DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission.J Mol Biol. 1998 Jun 26;279(5):1027-43. doi: 10.1006/jmbi.1998.1834. J Mol Biol. 1998. PMID: 9642081 Review.
Cited by
-
Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA.Nucleic Acids Res. 2010 Oct;38(19):6813-30. doi: 10.1093/nar/gkq521. Epub 2010 Jun 22. Nucleic Acids Res. 2010. PMID: 20571084 Free PMC article.
-
Regulation of rRNA transcription is remarkably robust: FIS compensates for altered nucleoside triphosphate sensing by mutant RNA polymerases at Escherichia coli rrn P1 promoters.J Bacteriol. 2000 Apr;182(7):1969-77. doi: 10.1128/JB.182.7.1969-1977.2000. J Bacteriol. 2000. PMID: 10715005 Free PMC article.
-
RNA polymerase and an activator form discrete subcomplexes in a transcription initiation complex.EMBO J. 2006 Aug 23;25(16):3784-90. doi: 10.1038/sj.emboj.7601261. Epub 2006 Aug 3. EMBO J. 2006. PMID: 16888625 Free PMC article.
-
Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli.J Bacteriol. 2001 Nov;183(21):6305-14. doi: 10.1128/JB.183.21.6305-6314.2001. J Bacteriol. 2001. PMID: 11591675 Free PMC article.
-
GeneWiz browser: An Interactive Tool for Visualizing Sequenced Chromosomes.Stand Genomic Sci. 2009 Sep 25;1(2):204-15. doi: 10.4056/sigs.28177. Stand Genomic Sci. 2009. PMID: 21304658 Free PMC article.
References
-
- Appleman J A. Ph.D. thesis. University of Wisconsin—Madison; 1998.
-
- Bartlett M S. Ph.D. thesis. University of Wisconsin—Madison; 1997.
-
- Bartlett M S, Gaal T, Ross W, Gourse R L. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrnP1 promoters. J Mol Biol. 1998;279:331–345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

